Abstract
Key message
EgMADS21 regulates PUFA accumulation in oil palm.
Abstract
Oil palm (Elaeis guineensis Jacq.) is the most productive world oil crop, accounting for 36% of world plant oil production. However, the molecular mechanism of the transcriptional regulation of fatty acid accumulation and lipid synthesis in the mesocarp of oil palm by up- or downregulating the expression of genes involved in related pathways remains largely unknown. Here, an oil palm MADS-box gene, EgMADS21, was screened in a yeast one-hybrid assay using the EgDGAT2 promoter sequence as bait. EgMADS21 is preferentially expressed in early mesocarp developmental stages in oil palm fruit and presents a negative correlation with EgDGAT2 expression. The direct binding of EgMADS21 to the EgDGAT2 promoter was confirmed by electrophoretic mobility shift assay. Subsequently, transient expression of EgMADS21 in oil palm protoplasts revealed that EgMADS21 not only binds to the EgDGAT2 promoter but also negatively regulates the expression of EgDGAT2. Furthermore, EgMADS21 was stably overexpressed in transgenic oil palm embryoids by Agrobacterium-mediated transformation. In three independent transgenic lines, EgDGAT2 expression was significantly suppressed by the expression of EgMADS21. The content of linoleic acid (C18:2) in the three transgenic embryoids was significantly decreased, while that of oleic acid (C18:1) was significantly increased. Combined with the substrate preference of EgDGAT2 identified in previous research, the results demonstrate the molecular mechanism by which EgMADS21 regulates EgDGAT2 expression and ultimately affects fatty acid accumulation in the mesocarp of oil palm.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The African oil palm (Elaeis guineensis Jacq.) is a perennial monocotyledonous plant belonging to the Arecaceae palm family and is the most productive oil-bearing crop in the world. Palm fruits have two storage tissues, the mesocarp and kernel, that can produce two oils of major economic importance, commonly referred to as palm oil and palm kernel oil (Teh et al. 2019). Palm oil, mainly referring to oil derived from the mesocarp of oil palm fruit, is in high demand due to its nutritional attributes and competitive price compared to other vegetable oils (Parveez et al. 2015a) The cultivation of oil palm has expanded greatly in recent years, such that it has now surpassed soybean oil as the most widely produced vegetable oil in the world (Loganathan et al.2017). Compared to other plant sources of oil, the major fatty acids (FAs) of palm oil are palmitic acid (16:0) and oleic acid (18:1) predominated (each about 40%), while only 8% of the FAs are polyunsaturated fatty acids (PUFAs) (Dussert et al. 2013). Although the role of palmitic acid as a potential cause of cardiovascular diseases is still a matter of debate, the production of less saturated oil with higher oleic and lower palmitic contents by genetic improvement remains one of the important objectives in the breeding of oil palm (Parveez et al. 2015b).
Diacylglycerol acyltransferases (DGATs) are a key group of enzymes that catalyze the final step in acyl-CoA-dependent triacylglycerol (TAG) biosynthesis, which has demonstrated it to be the important rate-limiting step of triacylglycerol production in plants (Routaboul et al. 1999; Zou et al. 1999). The first plant DGAT gene was isolated from Arabidopsis thaliana in 1999 (Zou et al. 1999), and then different types of DGATs have been identified from several kinds of species, including castor bean (Ricinus communis) (He et al. 2004), peanut (Arachis hypogea) (Zheng et al. 2017), and tung tree (Vernicia fordii) (Shockey et al. 2006). Current research works showed that there are four divergent types of DGAT (DGAT1, DGAT2, DGAT3 and WS/DGAT) in higher plants, sharing little homology with each other. For example, DGAT1 is the main enzyme responsible for TAG synthesis during seed development, while DGAT2s come from plant species that produce large amounts of unusual fatty acids (Iskandarov et al. 2017). In Arabidopsis, AtDGAT2 showed a different acyl-CoA substrate preference than AtDGAT1 (Zhou et al. 2013). Moreover, the DGAT3 was considered to be an enzyme participate in a cytosolic pathway of TAG biosynthesis (Hernandez et al. 2012), and WS/DGAT is believed to be responsible for formation of surface wax esters (Kalscheuer and Steinbuchel 2003).
In oil palm, four distinct functional families of DGAT enzymes have been identified, including DGAT1, DGAT2, DGAT3 and WS/DGAT (Rosli et al. 2018). Although metabolic control analysis to the Kennedy pathway for triacylglycerol formation in callus of oil palm indicated that DGAT is unlikely to affect oil accumulation in oil palm (Ramli et al. 2005), the evaluation of the gene expression of the four DGAT families in different tissues and developmental stages suggests that the four DGAT groups play distinct physiological roles in mesocarp and endosperm tissues (Rosli et al. 2018). In addition, one type of DGAT2 (XM_010933834) that is highly expressed in the mesocarp of oil palm was characterized for its in vivo activity. Seed-specific overexpression of EgDGAT2 in Arabidopsis thaliana increased the content of polyunsaturated 18:2 and 18:3 TAGs in the seeds compared to that in wild-type Arabidopsis. The results indicated that EgDGAT2 prefers to use polyunsaturated FAs as substrates in both yeast and plants (Jin et al. 2017). However, despite some progress in the gene cloning and functional analysis of the DGATs of oil palm, a more detailed understanding of the transcriptional regulation of DGATs in the mesocarp of oil palm is currently unavailable.
In plants, the MADS-box gene family is one of the best-studied gene families and plays important roles during plant development, especially in floral organ development (Airoldi and Davies 2012; Shan et al. 2009). The regulation of MADS-box genes contributes the majority of components in the classical ABC and expanded ABCE models of floral development (Cheng et al. 2017). Moreover, MADS-box genes have been found to be expressed in the endosperm, ovule, embryo and pericarp of fruit, suggesting their diverse roles in plant development (Kumar et al. 2016; Smaczniak et al. 2012). In oil palm, diverse MADs box transcripts were found to show expression associated with the different phases of mesocarp development in previous research. Comparative transcriptome analysis data indicate the involvement of MADS box genes, particularly AGAMOUS-like genes, during the maturation and ripening of the oil palm mesocarp (Tranbarger et al. 2011). However, it is still unknown whether there is a relationship between oil accumulation and transcriptional activity and whether these MADS box gene products play regulatory roles in fatty acid synthesis or TAG assembly during mesocarp ripening.
Here, we isolated EgMADS21, an AGAMOUS-like (AGL) MADS-box transcription factor (TF) that is highly expressed in the mesocarp of oil palm by yeast one-hybrid assay, and characterized its function in TAG biosynthesis by directly regulating the expression of the EgDGAT2 gene. In addition, the negative regulation of EgDGAT2 expression and reduced unsaturated fatty acid levels in transgenic oil palm cells were demonstrated. Finally, the potential for use of EgMADS21 in the genetic manipulation of oil accumulation in oil crops was discussed. In summary, we reveal a novel regulatory mechanism of EgMADS21-mediated TAG biosynthesis and oil accumulation in the mesocarp of oil palm.
Materials and methods
Plant materials and growth conditions
Oil palm (Elaeis guineensis Jacq.) fruits at the developmental stages of 30–60 days after pollination (DAP; Phase 1), 60–100 DAP (Phase 2), 100–120 DAP (Phase 3), 120–140 DAP (Phase 4) and 140–160 DAP (Phase 5) were harvested from the Coconut Research Institute, Chinese Agricultural Academy of Tropical Crops, Hainan, China (Zheng et al. 2019). The calli used in this study were induced from the young leaves of oil palm (Elaeis guineensis cv. Tenera). They were grown on MS medium supplemented with 5 mg/L 2, 4-D, 10 mg/L TDZ and 2% charcoal at 28 °C in the dark. The calli were transferred to woody plant medium (WPM) containing 0.1 mg/L 2, 4-D and 0.2% coconut water for secondary embryo induction and embryoid proliferation (Zou et al. 2019). The obtained embryoids were subcultured at 1-month intervals and used for genetic transformation and other assays.
DNA and RNA extraction and gene isolation
Genomic DNA was extracted based on the CTAB (cetyltrimethylammonium bromide) method. Total RNA was isolated by the CTAB method, and first-strand cDNA was synthesized from total RNA using TIANScript One Step RT-PCR Kits (Tiangen, Beijing, China). The primers for the EgDAGT2 promoter are listed in Supplemental Table 2. The PCR conditions were as follows: 98 °C for 30 s, 30 cycles of 98 °C for 10 s, 58.5 °C for 30 s, and 72 °C for 30 s, and a final step at 72 °C for 2 min.
Yeast one-hybrid assay
The Matchmaker™ One-Hybrid Library Screening system was used to identify proteins that bind to the EgDGAT2 promoter. The sequence of the EgDGAT2 promoter was cloned into the pHis yeast expression vector, which was then integrated into an oil palm one-hybrid library constructed with the pGADT7 AD vector in the Y187 yeast strain. The background histidine expression of the Y187 pHIS-EgDGAT2-promoter strain was tested and restrained with 3-amino-1, 2, 4-triazole (3-AT) at the appropriate concentration. Furthermore, the full length of each transcription factor obtained was cloned separately into the pGADT7 AD vector to confirm the screening results. The relationship between EgMADS21 and the EgDGAT2 promoter was examined individually.
Gene cloning and bioinformatics analysis
The open reading frame (ORF) cDNA sequence of EgMADS21 was cloned from oil palm mesocarp based on sequencing analysis using PCR. The products and original EST fragments were aligned and assembled into the full-length EgMADS21 cDNA (XM_010932108.2) using the DNAMAN program. The ORF Finder of NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to identify the ORF of EgMADS21.
The full-length of ORF nucleotide and amino acid sequence analyses were performed with the BLAST program from the NCBI website. Amino acid alignment of EgMADS21 was carried out with Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/), and a phylogenetic tree was constructed using the neighbor-joining method in MEGA 5.0 software (Tamura et al. 2007).
Real-time quantitative pcr (qRT-PCR) analysis
The cDNAs from oil palm mesocarp were diluted and mixed with primers and SYBR® Premix Ex Taq™II (TaKaRa, Dalian, China). qRT-PCR reactions were carried out in triplicate using SYBR® Premix Ex Taq™ II (TaKaRa, Tokyo, Japan) and were monitored with the CFX Connect™ Real-Time PCR Detection System (Bio-Rad, USA) according to the manufacturer’s instructions. The Egβ-actin housekeeping genes from oil palm were used as internal control genes. RT-PCR primers are listed in Supplemental Table 2. Expression levels were quantified as comparative cycle threshold (Ct) values using the 2−ΔΔCt method.
Subcellular localization analysis
The full-length cDNA of the EgMADS21 transcription factor was cloned into the p35MK1 vector. The construct was expressed transiently in tobacco (Nicotiana benthamiana) leaves by Agrobacterium tumefaciens-mediated infiltration (strain EHA105). The transiently infected leaves were imaged on day 2 after infiltration using a Leica (TCC-SP8) laser scanning confocal microscope. The 4′, 6-diamidino-2-phenylindole (DAPI) was used as a marker for nuclear localization. The excitation wavelength for GFP fluorescence was 488 nm, and fluorescence was detected at 490 to 520 nm.
Electrophoretic mobility shift assay (EMSA)
The protein expression vector pCold-EgMADS21 was constructed and transferred into the E. coli protein expression strain BL21. Mycoprotein production was induced by IPTG, and the product was obtained by ultrasonic processing. The recombinant protein was purified via Ni-chelating affinity chromatography, and the size and purity of the protein were verified by SDS-PAGE and Western blot analyses. Double-stranded segments were obtained by PCR, and a biotin label was added with a Biotin 3′ End DNA Labeling Kit (Thermo). The LightShift® Chemiluminescent EMSA Kit (Thermo) was used to perform EMSA experiments via the manufacturer’s instructions.
Vector construction for plant transformation
For transient and stable expression in oil palm, the ORF of EgMADS21 was amplified from cDNA using the 1300 s-EgMADS21 F/R primers and ligated into the binary expression vector pCAMBIA1300s under the control of the 35S promoter to generate the plant expression vector pCAMBIA1300-EgMADS21.
Transient expression of EgMADS21 in protoplasts
PEG-mediated transfection and analysis was performed as previously described (Jung et al. 2015) with minor modifications. Fresh oil palm leaves from young seedling were cut into 1 mm-wide threadlets and placed in a hydrolysate enzyme solution (containing cellulase, lyase, mannitol, KCl and MES) for 3 to 4 h. The protoplasts were collected by centrifugation at 100 g for 5 min, washed twice with washing solution and suspended in suspension buffer; the protoplasts could be stored temporarily at 4 °C. Then, the constructed vector was transferred to protoplasts via the PEG method. After the transfection, the protoplasts are collected by centrifugation for RNA extraction and RT-PCR. The expression levels of EgMADS21 and EgDGAT2 were analyzed by qRT-PCR as described above.
Stable expression of EgMADS21 in transgenic oil palm embryoids
The oil palm embryoids were subcultured under aseptic conditions and expanded by tissue culture. The MADS21-1300 s plasmid was electroporated into Agrobacterium EHA105, and the positive bacteria were screened by PCR. The embryoid was transformed by Agrobacterium infection according to the procedure described for Citrus sinensis callus (Li et al. 2003) with several modifications. The infected embryoids were grown on screening medium containing 30 mg/L hygromycin B (Sigma) to select positive transgenic embryoids. After 5 months of selection and subculture, the obtained transgenic embryoids were expanded and used for subsequent experiments (qRT-PCR and FAs analysis).
Total lipid extraction and fatty acid analysis
A chloroform–methanol (2:1 by volume) extraction solvent system was used for lipid extraction from the WT and transgenic embryoids of oil palm (Yuan et al. 2014). Dried samples were dissolved in 1 mL of n-hexane, and 1 mL of 14% BF3-methanol solvent was added for methyl esterification (Liang et al. 2014). The GC analysis of fatty acid methyl esters was performed on a Hewlett-Packard Ultra-GC instrument (Sun et al. 2017). An HP-FFAP capillary column (30 m × 0.25 mm, 0.25 μm thickness) was used to separate the fatty acid methyl esters.
Statistical analysis
Every sample was verified to show reproducibility using three biological replicates. Significant differences were determined by SPSS Statistics 19.0. Figures were prepared with GraphPad Prism 5. Student’s t test (*P < 0.05; **P < 0.01) was used to determine significant differences between two groups in this study.
Results
Screening of proteins interacting with the EgDGAT2 promoter
Firstly, based on genome blast, only one copy of EgDGAT2 was found on Elaeis guineensis genome (Chromosome 10). To identify transcription factors that regulate the lipid metabolic pathway by affecting the expression of EgDGAT2, a yeast one-hybrid library (Y187) screening system was applied in which the EgDGAT2 promoter was used as bait to screen an oil palm cDNA library. After screening, 54 different genes belonging to Elaeis guineensis were obtained based on sequence BLAST analysis (Supplemental Table 1). Among these genes, one clone encoding a MADS-box family protein (designated EgMADS21) was selected for analysis considering the important function of such genes in fruit development.
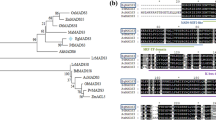
A yeast one-hybrid assay was performed to verify the interaction between EgMADS21 and the EgDGAT2 promoter. Cotransformation of fusion vectors (pHIS-p53-elements and pGADT7-Rec2-p53) that could combine in yeast Y187 was performed as a positive control. pGADT7-Rec2-p53 and pHIS without binding elements were used as negative controls. Bait yeast cells cotransformed with the fusion vector pGADT7-EgMADS21 could survive on the auxotrophic medium with 2 mM 3-AT (3-Amino-1, 2, 4-triazole). The growth of these colonies indicated that the EgMADS21 protein could interact with the EgDGAT2 promoter in the yeast system (Fig. 1a).
Screening of proteins interacting with the EgDGAT2 promoter and sequence analysis. a Yeast one-hybrid analysis of the interaction of EgMADS21 and the EgDGAT2 promoter. pHIS-p53-elements and pGADT7-Rec2-p53 were used as the positive control, and pHIS and pGADT7-Rec2-p53 were used as the negative control. SD-His/Leu/Trp, SD medium without His, Leu, Trp, or Leu supplemented with 3-AT at a concentration of 2 mM. b Multiple sequence alignment of EgMADS21 and related proteins from Arabidopsis and palm plants. The horizontal lines mark the four conserved domains. c Phylogenetic analysis of EgMADS21 and related proteins from other plant species. The scale bar represents 0.1 substitutions per site. The protein sequences include EgMADS21 (XP_010930410), EgMADS2 (XP_010911265) and EgMADS9 (NP_001306837) from Elaeis guineensis, AtMADS (CAA19810.1) from Arabidopsis thaliana, PdMADS (XP_008781978) from Phoenix dactylifera, CnMADS (AIK66813) from Cocos nucifera, OsMADS (AAK17066) from Oryza sativa, TaMADS (AAQ11687) from Triticum aestivum, GmMADS (NP_001236390) from Glycine max, ZmMADS (NP_001105692) from Zea mays, StMADS (NP_001274754) from Solanum tuberosum and AvMADS (BAD83772) from Asparagus virgatus
Gene cloning, sequence analysis and expression of EgMADS21
The open reading frame (ORF) cDNA sequence (708 bp) of EgMADS21 was cloned from oil palm mesocarp based on sequencing analysis using PCR. Bioinformatics analysis indicated that EgMADS21 contains 708 nucleotides, which encode 235 amino acids containing four typical domains: a MADS-box domain, a variable I domain, a conserved K domain, and a C-terminal region (Fig. 1b). Phylogenetic analysis showed that EgMADS21 indeed belongs to the AGAMOUS (AG)-like group. In addition, three typical MADS sequences from oil palm (EgMADS2, EgMADS21 and EgMADS9) were grouped into two subclades; EgMADS21 was closely linked to PdMADS, CnMADS and ApMADS, whereas EgMADS2 and EgMADS9 were grouped with plants that do not belong to Palmae, such as Arabidopsis and Oryza sativa (Fig. 1c).
Expression pattern of EgMADS21 and its subcellular localization
To understand the dynamic relationship between the EgMADS21 and EgDGAT2 genes during fruit development in oil palm, qRT-PCR was applied to analyze the expression levels of the two genes in five different stages of fruit development. The results indicated differences in their expression levels at five developmental stages (Fig. 2a); in the first three stages of oil palm fruit development, EgDGAT2 expression was very stable. However, the expression level increased significantly during the fourth and fifth stages of fruit ripening. In contrast, EgMADS21 exhibited higher expression levels in the first three stages of fruit development and lower expression levels in the last two stages of fruit ripening. The expression level of EgDGAT2 was negatively correlated with that of EgMADS21, suggesting that EgMADS21 may negatively regulate the expression of EgDGAT2 during the development of oil palm fruits. At the same time, according to the dynamic change of fatty acids in the mesocarp of oil palm in the previous references (Dussert et al. 2013; Zheng et al. 2019), the expression of EgMADS21 is also negatively correlated with the increase of unsaturated fatty acids in this process.
To elucidate the subcellular localization pattern of EgMADS21 in plant cells, the CDS region (in which stop codon was deleted) was inserted into the p35MK1 vector to construct an EgMADS21-GFP fusion gene, which was then transiently expressed by infiltration in tobacco leaves. As shown in Fig. 2b, the localization results showed that EgMADS21 localized to the nucleus.
Verification of the interaction of EgMADS21 and the EgDGAT2 promoter
To confirm the binding of EgMADS21 at the EgDGAT2 promoter, EMSA was performed using prokaryon-expressed and purified EgMADS21-His fusion proteins. The binding site of the EgDGAT2 promoter was predicted and designed with AliBaba2.1 (http://gene-regulation.com/pub/programs/alibaba2/index.html) and 5 potential binding sites were predicted (Supplemental Fig. 1). The 50 bp fragments containing the binding site was amplified by PCR; the biotin tag was added as a probe; and a fragment without a biotin tag was added as a competitive sequence. After EMSA verification, only one site was proved can bind with EgMADS21. In the presence of the inhibitor, it can be seen from the EMSA results that the protein expressed from pCOLD without a label did not bind to the promoter fragment, whereas the expressed EgMADS21 protein showed a band corresponding to binding with the probe, confirming that EgMADS21 can bind to the element in this fragment of the EgDGAT2 promoter (Fig. 3a).
EMSA and transient expression of EgMADS21 in the protoplasts of oil palm. a Binding of EgMADS21 to the motif in the promoter of the EgDGAT2 gene. Wild protein means the protein expressed from plasmid pCOLD without EgMADS21. b Transient overexpression of EgMADS21 in the protoplasts of oil palm. 1300S: Empty vector (control)
Transient expression of EgMADS21 in protoplasts
Protoplasts were extracted from wild-type oil palm leaves, and the pCAMBIA1300s (as blank) and pCAMBIA1300-EgMADS21 plant expression vectors were transferred to fresh oil palm protoplasts via the PEG induction method, from which RNA was extracted after overnight transformation. According to the results of fluorescence quantification (Fig. 3b), the expression level of EgMADS21 was significantly increased, indicating that the pCAMBIA1300-EgMADS21 plasmid was successfully transferred, while the expression of EgDGAT2 was significantly decreased. Combined with the EMSA results, it was shown that EgMADS21 not only binds to the EgDGAT2 promoter but also negatively regulates EgDGAT2 expression.
Overexpression of EgMADS21 in Oil palm embryoids
To further verify the influence of EgMADS21 on EgDGAT2, EgMASD21 was stably overexpressed in the embryoids of Elaeis guineensis. After three rounds of hygromycin screening and PCR identification, three positive transgenic lines of embryoids were selected for further analysis (Fig. 4a). The relative expression levels of EgMADS21 and EgDGAT2 in wild and transgenic embryoids were quantified by qRT-PCR, and it was found that EgMADS21 transcript expression in the embryoids varied up to sixfold and 33-fold, respectively, compared to that in wild embryoids (Supplement Table 3). In contrast, the expression of EgDGAT2 declined remarkably in the EgMADS21-2 line, and EgDGAT2 expression was suppressed substantially (Fig. 4b). In conclusion, EgDGAT2 expression was seriously inhibited, while the expression of EgMADS21 was increased.
Stable overexpression of EgMADS21 in the embryoids of oil palm and fatty acid analysis. a Wild-type and transgenic oil palm embryoids after selection. b Expression levels of EgMADS21 in different transgenic lines. c Total FA content in different transgenic lines. d FA profiling of different transgenic lines
Fatty acid analysis
To investigate the effects of the overexpression of EgMADS21 in transgenic embryoids, we extracted the total lipids from three individual transformation-positive lines and wild-type embryoids and analyzed the contents of different fatty acids by GC. The results showed that the contents of C18:1, C20:0, and C20:1 in the three transgenic callus lines were increased significantly compared with the wild-type calli. More specifically, the EgMADS21 transgenic embryoids contained 23.1 mol % C18:1 FA on average, while the content was only 15.53 mol % in WT embryoids. In addition, the content of 18:2 FA in the transgenic embryoids of EgMADS21-overexpressing lines was decreased from 32.5 to 23.6 mol % compared with that in WT. However, compared with the WT, the contents of linoleic acid (C18:2) in the three types of transgenic embryoids were obviously decreased. Moreover, the contents of long-chain fatty acids, including C20:0, C20:1, C22:1 and C24:0, were increased significantly (Fig. 4c). Therefore, combined with the substrate preference of EgDGAT2 identified in earlier research (Jin et al. 2017), the results of fatty acid analysis further demonstrated that EgMADS21 inhibits the expression of the EgDGAT2 gene by directly interacting with the DGAT2 promoter and ultimately decreases polyunsaturated fatty acid accumulation during TAG synthesis in oil palm.
Discussion
In this study, we described an AGAMOUS-like MADS box TF, EgMADS21, that modulates unsaturated FAs during TAG accumulation via the regulation of key metabolic gene. EgMADS21 was identified as an inhibitory factor of the EgDGAT2 gene promoter, which is involved in a key step in TAG synthesis during mesocarp maturation. In Arabidopsis thaliana, members of the AGAMOUS subfamily of MADS-box genes have been demonstrated to control ovule development and were subsequently revealed to participate in the transition to flowering (Hugouvieux et al. 2018; Koo et al. 2010; Zheng et al. 2009). Furthermore, Sl-AGL11 from tomato is involved in the conversion of sepals into fleshy organs under ethylene-dependent ripening and starch and sugar accumulation during fruit ripening (Huang et al. 2017). MaMADS7 from banana (Musa acuminata) plays an important role in initiating endogenous ethylene biosynthesis and fruit ripening (Liu et al. 2015). In the mesocarp of oil palm, the expression of the EgMADS21 gene is high in the early fruit stages (60–100 DAP) and decreases gradually with fruit ripening. In contrast, the expression level of EgDGAT2 does not change during the early developmental stages and increases significantly with fruit ripening (Jin et al. 2017). Correlated with changes in EgDGAT2 expression, the polyunsaturated fatty acid (PUFA) content dramatically declines during this period. Considering that EgDGAT2 preferentially uses PUFAs (especially C18:3 and C18:3 PUFAs) as substrates in transgenic plants, EgMADS21 may modulate TAG assembly and PUFA accumulation by suppressing EgDGAT2 expression during the ripening of oil palm fruit (Fig. 5).
Simplified model shows that EgMADS21 directly and indirectly regulates the expression of the EgDGAT2 gene, which controls the accumulation of fatty acids in oil palm. DAG diacylglycerol, G3P glycerol 3-phosphate, GPAT glycerol-3-phosphate acyltransferase, LPA lysophosphatidic acid, LPAAT lysophosphatidic acid acyltransferase, PA phosphatidic acid, TAG Triacylglycerols
In higher plants, the complicated networks of seed oil accumulation are precisely controlled by intricate multilevel regulatory networks, among which multiple routes are regulated in a coordinated fashion, including carbon partitioning, FA synthesis, and lipid assembly (Tan et al. 2019). Previous transcriptomic profiling analyses of different kinds of plant species also indicated that transcriptional regulation plays an important role in the lipid biosynthesis process (Chen et al. 2012; Liang et al. 2014; Lu et al. 2018). Several kinds of transcription factors (TFs), including WRINKLED1 (WRI1), LEAFY COTYLEDON1 (LEC1), LEC2, FUSCA3 (FUS3), and an R2R3-MYB (MYB89) transcription factor, were identified in these analyses, and the regulatory network of the TFs controlling seed oil accumulation was revealed (Baud et al. 2007; Kagaya et al. 2005; Roscoe et al. 2018). Among these TFs, WRI1, belonging to the APETALA2-ethylene-responsive element-binding protein family, was demonstrated to be a central regulator in seed oil accumulation by interacting directly with an AWbox sequence, [CnTnG(n)7CG], in the upstream regions of several FA biosynthesis genes during seed maturation (Maeo et al. 2009). In addition, in the seeds of Arabidopsis thaliana, WRI1 and FUS3 are positively regulated by LEC1 and LEC2 and jointly control the expression of genes related to the accumulation of seed oil (Baud et al. 2007; Kong and Ma 2018; Maeo et al. 2009). In oil palm, WRI1-like transcript levels are high in oil palm mesocarp and continuously increase during ripening (Dussert et al. 2013). However, in contrast to what is observed in Arabidopsis thaliana, EgWRI1-1 in the mesocarp of oil palm is activated by three transcription factors, EgNF-YA3, EgNF-YC2 and EgABI5, as no homologs of LEC1 and 2 and FUS3 were identified in oil palm mesocarp (Yeap et al. 2017). The results suggest that TFs are likely to control oil accumulation and FA synthesis in the mesocarp of oil palm by different regulatory networks, which are independent of the upstream factors that participate in seed development.
Most TFs related to oil synthesis are derived from oil crop seeds in which TAGs accumulate during maturation (Baud et al. 2010; Baud and Lepiniec 2010; Li et al. 2015). In contrast, little is known about the molecular basis of the regulation of lipid metabolism by TFs in fleshy fruits that accumulate high levels of TAGs in their mesocarp, such as those of oil palm and avocado (Persea americana). The fruit-specific regulatory network appears to be controlled by ethylene-dependent and abscisic acid (ABA)-dependent pathways and other independent pathways (Kourmpetli and Drea 2014). The members of the MADS box family of transcription factors (TFs), including the class D MADS-box gene Sl-AGL11 from tomato (Huang et al. 2017), the MdAGL24-like gene in red-fleshed apple (Malus sieversii f. niedzwetzkyana) (Su et al. 2018), and the TM6 MADS-box gene from strawberry (Fragaria × ananassa) (Martin-Pizarro et al. 2019), have been identified as common regulatory elements during fruit ripening. However, possibly due to a lack of data, only a few MADS box family TFs have been identified as being involved in FA biosynthesis during maturation and ripening of oil palm fruit, which presents an especially abundant oil content in its mesocarp (Yeap et al. 2017). During the maturation and ripening of the oil palm mesocarp, the expression of certain MADS box transcripts coincides with oil accumulation and FA synthesis in a positive and negative manner, respectively (Tranbarger et al. 2011). These results support the hypothesis that some MADS box proteins play regulatory roles either upstream or downstream of oil accumulation or FA synthesis during mesocarp ripening.
Although a large number of contigs with differential representation have been revealed by comparative transcriptome and metabolite analysis of the oil palm mesocarp, very few studies have focused on the MADS-box TFs involved in the regulatory mechanism underlying FA synthesis and TAG assembly during mesocarp development (Dussert et al. 2013). The selection of a suitable model plant for the functional analysis of MADS-box TFs derived from oil palm has caused a dilemma for researchers. Although knowledge obtained from the model species Arabidopsis has advanced enough to characterize oil accumulation-related TFs from nonmodel plant species (Yeap et al. 2017), whether the functions of these TFs from mesocarp (nonseed tissues) are conserved and play a role in seed oil synthesis in Arabidopsis remains unclear. In this kind of tissue, the ripening process is always closely controlled by both ethylene-dependent and ethylene-independent pathways (Gapper et al. 2014; Giovannoni 2007). While tomato is a good model plant for examining fleshy fruit development and metabolite accumulation (Dong et al. 2013), its low content or lack of triacylglycerols (TAGs) means that it is not an optimal model for studying the transcriptional regulation of downstream pathways of FAs and TAG synthesis (Tranbarger et al. 2011). Moreover, the elucidation of MADS box-mediated regulation might be particularly challenging in the fruit of oil palm, because MADS-box genes are known to have redundant functions, and TFs may be involved in multiple pathways; hence, their ectopic expression may result in pleiotropic phenotypes (Zhang 2003). Therefore, functional characterization of the components involved in oil synthesis via embryogenic callus transformation combined with virus-induced gene silencing (VIGS) (Wang and Fu 2018) in fleshly fruit will be an alternative method for future research.
Based on the above results and discussion, EgMADS21 was characterized as a regulator of TAG metabolism in this study. As shown in Fig. 5, the underlying molecular pathway responsible for the effects of EgMADS21 involves inhibition of EgDGAT2 expression and a decrease in PUFAs assembled at the 3-n of DAG, finally leading to decreased PUFA accumulation in the mesocarp of oil palm. To our knowledge, this is the first report to clearly demonstrate the mechanism by which a MADS-box family gene of oil palm negatively regulates fatty acid accumulation during fruit development. The results also provide new insight into the potential functions played by MADS-box family genes in transcriptional regulatory mechanisms, such as the qualitative and quantitative control of oil palm fruit development and metabolism.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Airoldi CA, Davies B (2012) Gene duplication and the evolution of plant MADS-box transcription factors. J Genet Genomics 39:157–165
Baud S, Lepiniec L (2010) Physiological and developmental regulation of seed oil production. Prog Lipid Res 49:235–249
Baud S, Mendoza MS, To A, Harscoet E, Lepiniec L, Dubreucq B (2007) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J 50:825–838
Baud S, Feria Bourrellier AB, Azzopardi M, Berger A, Dechorgnat J, Daniel-Vedele F, Lepiniec L, Miquel M, Rochat C, Hodges M, Ferrario-Mery S (2010) PII is induced by WRINKLED1 and fine-tunes fatty acid composition in seeds of Arabidopsis thaliana. Plant J 64:291–303
Chen H, Wang FW, Dong YY, Wang N, Sun YP, Li XY, Liu L, Fan XD, Yin HL, Jing YY, Zhang XY, Li YL, Chen G, Li HY (2012) Sequence mining and transcript profiling to explore differentially expressed genes associated with lipid biosynthesis during soybean seed development. BMC Plant Biol 12:122
Cheng Z, Ge W, Li L, Hou D, Ma Y, Liu J, Bai Q, Li X, Mu S, Gao J (2017) Analysis of MADS-Box gene family reveals conservation in floral organ ABCDE model of Moso bamboo (Phyllostachys edulis). Front Plant Sci 8:656
Dong T, Hu Z, Deng L, Wang Y, Zhu M, Zhang J, Chen G (2013) A tomato MADS-Box transcription factor, SlMADS1, acts as a negative regulator of fruit ripening. Plant Physiol 163:1026–1036
Dussert S, Guerin C, Andersson M, Joet T, Tranbarger TJ, Pizot M, Sarah G, Omore A, Durand-Gasselin T, Morcillo F (2013) Comparative transcriptome analysis of three oil palm fruit and seed tissues that differ in oil content and fatty acid composition. Plant Physiol 162:1337–1358
Gapper NE, Giovannoni JJ, Watkins CB (2014) Understanding development and ripening of fruit crops in an ‘omics’ era. Hortic Res 1:14034
Giovannoni JJ (2007) Fruit ripening mutants yield insights into ripening control. Curr Opin Plant Biol 10:283–289
He X, Chen GQ, Lin JT, McKeon TA (2004) Regulation of diacylglycerol acyltransferase in developing seeds of castor. Lipids 39:865–871
Hernandez ML, Whitehead L, He ZS, Gazda V, Gilday A, Kozhevnikova E, Vaistij FE, Larson TR, Graham IA (2012) A cytosolic acyltransferase contributes to triacylglycerol synthesis in sucrose-rescued arabidopsis seed oil catabolism mutants. Plant Physiol 160:215–225
Huang B, Routaboul JM, Liu M, Deng W, Maza E, Mila I, Hu G, Zouine M, Frasse P, Vrebalov JT, Giovannoni JJ, Li Z, van der Rest B, Bouzayen M (2017) Overexpression of the class D MADS-box gene Sl-AGL11 impacts fleshy tissue differentiation and structure in tomato fruits. J Exp Bot 68:4869–4884
Hugouvieux V, Silva CS, Jourdain A, Stigliani A, Charras Q, Conn V, Conn SJ, Carles CC, Parcy F, Zubieta C (2018) Tetramerization of MADS family transcription factors SEPALLATA3 and AGAMOUS is required for floral meristem determinacy in Arabidopsis. Nucleic Acids Res 46:4966–4977
Iskandarov U, Silva JE, Kim HJ, Andersson M, Cahoon RE, Mockaitis K, Cahoon EB (2017) A specialized diacylglycerol acyltransferase contributes to the extreme medium-chain fatty acid content of cuphea seed oil. Plant Physiol 174:97–109
Jin Y, Yuan Y, Gao L, Sun R, Chen L, Li D, Zheng Y (2017) Characterization and functional analysis of a type 2 diacylglycerol acyltransferase (DGAT2) gene from oil palm (Elaeis guineensis Jacq.) Mesocarp in Saccharomyces cerevisiae and transgenic Arabidopsis thaliana. Front Plant Sci 17(8):1791
Jung HI, Yan J, Zhai Z, Vatamaniuk OK (2015) Gene functional analysis using protoplast transient assays. Methods Mol Biol 1284:433–452
Kagaya Y, Toyoshima R, Okuda R, Usui H, Yamamoto A, Hattori T (2005) LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol 46:399–406
Kalscheuer R, Steinbuchel A (2003) A novel bifunctional wax ester synthase/acyl-CoA:diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. J Biol Chem 278:8075–8082
Kong Q, Ma W (2018) WRINKLED1 transcription factor: How much do we know about its regulatory mechanism? Plant Sci 272:153–156
Koo SC, Bracko O, Park MS, Schwab R, Chun HJ, Park KM, Seo JS, Grbic V, Balasubramanian S, Schmid M, Godard F, Yun DJ, Lee SY, Cho MJ, Weigel D, Kim MC (2010) Control of lateral organ development and flowering time by the Arabidopsis thaliana MADS-box Gene AGAMOUS-LIKE6. Plant J 62:807–816
Kourmpetli S, Drea S (2014) The fruit, the whole fruit, and everything about the fruit. J Exp Bot 65:4491–4503
Kumar G, Arya P, Gupta K, Randhawa V, Acharya V, Singh AK (2016) Comparative phylogenetic analysis and transcriptional profiling of MADS-box gene family identified DAM and FLC-like genes in apple (Malusx domestica). Sci Rep 6:20695
Li DD, Shi W, Deng XX (2003) Factors influencing Agrobacterium-mediated embryogenic callus transformation of Valencia sweet orange (Citrus sinensis) containing the pTA29-barnase gene. Tree Physiol 23:1209–1215
Li F, Wang W, Zhao N, Xiao B, Cao P, Wu X, Ye C, Shen E, Qiu J, Zhu QH, Xie J, Zhou X, Fan L (2015) Regulation of nicotine biosynthesis by an endogenous target mimicry of MicroRNA in tobacco. Plant Physiol 169:1062–1071
Liang Y, Yuan Y, Liu T, Mao W, Zheng Y, Li D (2014) Identification and computational annotation of genes differentially expressed in pulp development of Cocos nucifera L. by suppression subtractive hybridization. BMC Plant Biol 14(1):205
Liu J, Liu L, Li Y, Jia C, Zhang J, Miao H, Hu W, Wang Z, Xu B, Jin Z (2015) Role for the banana AGAMOUS-like gene MaMADS7 in regulation of fruit ripening and quality. Physiol Plant 155:217–231
Loganathan R, Subramaniam KM, Radhakrishnan AK, Choo YM, Teng KT (2017) Health-promoting effects of red palm oil: evidence from animal and human studies. Nutr Rev 75:98–113
Lu S, Sturtevant D, Aziz M, Jin C, Li Q, Chapman KD, Guo L (2018) Spatial analysis of lipid metabolites and expressed genes reveals tissue-specific heterogeneity of lipid metabolism in high- and low-oil Brassica napus L. seeds. Plant J 94:915–932
Maeo K, Tokuda T, Ayame A, Mitsui N, Kawai T, Tsukagoshi H, Ishiguro S, Nakamura K (2009) An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J 60:476–487
Martin-Pizarro C, Trivino JC, Pose D (2019) Functional analysis of the TM6 MADS-box gene in the octoploid strawberry by CRISPR/Cas9-directed mutagenesis. J Exp Bot 70:885–895
Parveez GK, Bahariah B, Ayub NH, Masani MY, Rasid OA, Tarmizi AH, Ishak Z (2015a) Production of polyhydroxybutyrate in oil palm (Elaeis guineensis Jacq.) mediated by microprojectile bombardment of PHB biosynthesis genes into embryogenic calli. Plant Sci 6:598
Parveez GK, Rasid OA, Masani MY, Sambanthamurthi R (2015b) Biotechnology of oil palm: strategies towards manipulation of lipid content and composition. Plant Cell Rep 34:533–543
Ramli US, Salas JJ, Quant PA, Harwood JL (2005) Metabolic control analysis reveals an important role for diacylglycerol acyltransferase in olive but not in oil palm lipid accumulation. FEBS J 272:5764–5770
Roscoe TJ, Vaissayre V, Paszkiewicz G, Clavijo F, Kelemen Z, Michaud C, Lepiniec L, Dubreucq B, Zhou DX, Devic M (2018) Regulation of FUSCA3 expression during seed development in Arabidopsis. Plant Cell Physiol 60(2):476–487
Rosli R, Chan PL, Chan KL, Amiruddin N, Low EL, Singh R, Harwood JL, Murphy DJ (2018) In silico characterization and expression profiling of the diacylglycerol acyltransferase gene family (DGAT1, DGAT2, DGAT3 and WS/DGAT) from oil palm, Elaeis guineensis. Plant Sci 275:84–96
Routaboul JM, Benning C, Bechtold N, Caboche M, Lepiniec L (1999) The TAG1 locus of Arabidopsis encodes for a diacylglycerol acyltransferase. Plant Physiol Biochem 37:831–840
Shan H, Zahn L, Guindon S, Wall PK, Kong H, Ma H, DePamphilis CW, Leebens-Mack J (2009) Evolution of plant MADS box transcription factors: evidence for shifts in selection associated with early angiosperm diversification and concerted gene duplications. Mol Biol Evol 26:2229–2244
Shockey JM, Gidda SK, Chapital DC, Kuan JC, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM (2006) Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18:2294–2313
Smaczniak C, Immink RG, Angenent GC, Kaufmann K (2012) Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139:3081–3098
Su M, Wang N, Jiang S, Fang H, Xu H, Wang Y, Zhang Z, Zhang J, Xu L, Zhang Z, Chen X (2018) Molecular characterization and expression analysis of the critical floral gene MdAGL24-like in red-fleshed apple. Plant Sci 276:189–198
Sun R, Ye R, Gao L, Zhang L, Wang R, Mao T, Zheng Y, Li D, Lin Y (2017) Characterization and ectopic expression of CoWRI1, an AP2/EREBP domain-containing transcription factor from coconut (Cocos nucifera L.) endosperm, changes the seeds oil content in transgenic Arabidopsis thaliana and Rice (Oryza sativa L. Front Plant Sci 8:63
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tan H, Zhang J, Qi X, Shi X, Zhou J, Wang X, Xiang X (2019) Correlation analysis of the transcriptome and metabolome reveals the regulatory network for lipid synthesis in developing Brassica napus embryos. Plant Mol Biol 99:31–44
Teh CK, Lee HL, Abidin H, Ong AL, Mayes S, Chew FT, Appleton D (2019) A practical genome-enabled legitimacy assay for oil palm breeding and seed production. BMC Plant Biol 19:470
Tranbarger TJ, Dussert S, Joet T, Argout X, Summo M, Champion A, Cros D, Omore A, Nouy B, Morcillo F (2011) Regulatory mechanisms underlying oil palm fruit mesocarp maturation, ripening, and functional specialization in lipid and carotenoid metabolism. Plant Physiol 156:564–584
Wang C, Fu D (2018) Virus-induced gene silencing of the eggplant chalcone synthase gene during fruit ripening modifies epidermal cells and gravitropism. J Agric Food Chem 66:2623–2629
Yeap WC, Lee FC, Shabari Shan DK, Musa H, Appleton DR, Kulaveerasingam H (2017) WRI1-1, ABI5, NF-YA3 and NF-YC2 increase oil biosynthesis in coordination with hormonal signaling during fruit development in oil palm. Plant J 91:97–113
Yuan YJ, Chen YH, Yan S, Liang YX, Zheng YS, Li DD (2014) Molecular cloning and characterisation of an acyl carrier protein thioesterase gene (CocoFatB1) expressed in the endosperm of coconut (Cocos nucifera) and its heterologous expression in Nicotiana tabacum to engineer the accumulation of different fatty acids. Funct Plant Biol 41:80–86
Zhang JZ (2003) Overexpression analysis of plant transcription factors. Curr Opin Plant Biol 6:430–440
Zheng Y, Ren N, Wang H, Stromberg AJ, Perry SE (2009) Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21:2563–2577
Zheng L, Shockey J, Bian F, Chen G, Shan L, Li X, Wan S, Peng Z (2017) Variant amino acid residues alter the enzyme activity of peanut type 2 diacylglycerol acyltransferases. Front Plant Sci 8:1751
Zheng Y, Chen C, Liang Y, Sun R, Gao L, Liu T, Li D (2019) Genome-wide association analysis of the lipid and fatty acid metabolism regulatory network in the mesocarp of oil palm (Elaeis guineensis Jacq.) based on small noncoding RNA sequencing. Tree Physiol 39:356–371
Zhou XR, Shrestha P, Yin F, Petrie JR, Singh SP (2013) AtDGAT2 is a functional acyl-CoA:diacylglycerol acyltransferase and displays different acyl-CoA substrate preferences than AtDGAT1. FEBS Lett 587:2371–2376
Zou J, Wei Y, Jako C, Kumar A, Selvaraj G, Taylor DC (1999) The Arabidopsis thaliana TAG1 mutant has a mutation in a diacylglycerol acyltransferase gene. Plant J 19:645–653
Zou JX, Zhang Q, Zhu ZY, Gao LC, Zheng YS, Li DD (2019) Embryogenic callus induction and fatty acid composition analysis of oil palm (Elaeis guineensis cv. Tenera). Sci Hortic-Amsterdam 245:125–130
Acknowledgements
We are grateful to the following investigators for helping with the tissue culture and fatty acid analysis: Ms. Li Gao and Mr. Lizhi Chen at Hainan University.
Funding
This research was supported by the Hainan Provincial Natural Science Foundation of China (No. 2019CXTD397), the National Natural Science Foundation of China (NSFC) (No. 31460213 and 31660222), National Key R&D Program of China,2018YFD1000500 and the Fundamental Research Funds for Chinese Academy of Tropical Agricultural Sciences (No.1630052019001).
Author information
Authors and Affiliations
Contributions
DL and YZ contributed substantially to the experimental design, conceived of the study and revised the article. SL, QZ and YJ carried out mainly experiments, drafted the manuscript, comprehensively analyzed data from all experimental results. DL gave substantial suggestions to the paper writing and language organization. JZ performed the mainly embryoids culture and transformation. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Günther Hahne.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Sy., Zhang, Q., Jin, Yh. et al. A MADS-box gene, EgMADS21, negatively regulates EgDGAT2 expression and decreases polyunsaturated fatty acid accumulation in oil palm (Elaeis guineensis Jacq.). Plant Cell Rep 39, 1505–1516 (2020). https://doi.org/10.1007/s00299-020-02579-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-020-02579-z