Abstract
Cyclins, cyclin-dependent kinases, and a number of other proteins control the progression of plant cell cycle. Although extensive studies have revealed the roles of some cell cycle regulators and the underlying mechanisms in Arabidopsis, relatively a small number of cell cycle regulators were functionally analyzed in rice. In this study, we describe 41 regulators in the rice genome. Our results indicate that the rice genome contains a less number of the core cell cycle regulators than the Arabidopsis one does, although the rice genome is much larger than the Arabidopsis one. Eight groups of CDKs similar to those in Arabidopsis were identified in the rice genome through phylogenetic analysis, and the corresponding members in the different groups include E2F, CKI, Rb, CKS and Wee. The structures of the core cell regulators were relatively conserved between the rice and Arabidopsis genomes. Furthermore, the expression of the majority of the core cell cycle genes was spatially regulated, and the most closely related ones showed very similar patterns of expression, suggesting functional redundancy and conservation between the highly similar core cell cycle genes in rice and Arabidopsis. Following auxin or cytokinin treatment, the expression of the core cell cycle genes was either upregulated or downregulated, suggesting that auxin and/or cytokinin may directly regulate the expression of the core cell cycle genes. Our results provide basic information to understand the mechanism of cell cycle regulation and the functions of the rice cell cycle genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Progression of cell cycle is primarily controlled by a highly conserved molecular machinery in which cyclin-dependent kinases (CDKs) play a central role. CDK activity is regulated in a complex manner, including reversible phosphorylation by specific kinases/phosphatases and association with other regulatory proteins. Core cell cycle regulators, such as CDKs, cyclins, CDK inhibitors (CKIs), homologs of the retinoblastoma protein (Rb) and the E2F transcription factors (E2F), have been identified in eukaryotes. In higher plants, the cell cycle must be integrated into a complex system of histogenesis and organogenesis since the spatial and temporal regulation of cell division is essential to the development of plant form (Mironov et al. 1997; Meijer and Murray 2001; Stals and Inzé 2001; De Veylder et al. 2003).
A large number of core cell cycle genes have been identified in many plant species. In Arabidopsis, 99 cell cycle genes have been classified into 7 families using a homology-based annotation protocol, and CDKs and cyclins are two large families (Potuschak and Doerner 2001; Vandepoele et al. 2002; Menges et al. 2005). Based on phylogenetic analysis, 8 classes of CDKs (CDKA-G, CDK-LIKE, CKL) and 10 classes of cyclins (A, B, C, D, H, L, T, U, SDS, and J18) were further defined in Arabidopsis (Vandepoele et al. 2002; Wang et al. 2004; Menges et al. 2005). Most of the core cell cycle genes were expressed across almost all tissues at detectable levels in the cell suspension culture, and a model has been proposed for a transcriptional regulation of the plant cell cycle (Beemster et al. 2005; Menges et al. 2002, 2003, 2005). The cell cycle is influenced by extracellular cues as well. Plant hormones are important in the regulation of the cell cycle. Of them, both cytokinins and auxins have been extensively documented for their roles in controlling the transcription of the cell cycle genes (Frank and Schmülling, 1999; Rashotte et al. 2003; Roudier et al. 2003; Pozo et al. 2005; Wang et al. 2006).
Several core cell cycle genes, such as RBR1, RBR2 and RBR3, were isolated from maize, and their functions were analyzed (Grafi et al. 1996; Ach et al. 1997; Hsieh and Wolniak 1998; Sabelli et al. 2005; Sabelli and Larkins 2006). In rice, the expression patterns of four CDK genes were examined (Umeda et al. 1999), and OsCKI1 had a role in root development and responded to both brassinosteroid and abscisic acid (Liu et al. 2003). Further analysis of the cyclin gene family in rice was conducted phylogenetically (La et al. 2006). Although extensive and systematic studies on the functions of core cell cycle genes have been reported in Arabidopsis, only a small number of core cell cycle genes were functionally examined in monocots. Rice, as a model plant species of monocots, is suitable to the identification of all core cell cycle genes because its whole genome sequence is available. In this study, we searched for the predicated core cell cycle genes from rice genome and conducted phylogenetic analysis based on their protein sequences using Arabidopsis queries (Vandepoele et al. 2002; Wang et al. 2004). Furthermore, we examined their expression patterns in plant tissues and the regulation of their expression by cytokinin and auxin. Our results will aid our understanding on the mechanism of cell cycle regulation and the functions of core cell cycle genes in rice.
Materials and methods
Plant materials
Rice plants (Orysa sativa L. cv. Zhong hua 11, obtained from Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai) were grown under natural growth conditions in the field at the Agricultural Experiment Station of Shandong Agricultural University.
To investigate the responses of the core cell cycle genes to hormones, roots of 2 week-old rice seedlings were submerged in a solution of either 10 μM IAA or 5 μM 6-BA, and the seedlings were then harvested in times as indicated for mRNA isolation.
Data retrieval and phylogenetic analysis
The cyclin-like proteins in rice were searched by using FGP-MINE Programs on the Floral Genome Project Website (http://www.fgp.bio.psu.edu/cgi-bin/fgpmine/fgp_family_list.cgi) (Wang et al. 2004). To identify other core cell cycle proteins in rice, BLAST searches and sequence alignments were performed against the KOME (http://www.cdna01.dna.affrc.go.jp/cDNA/) and NCBI (http://www.ncbi.nlm.nih.gov/) databases. We first used the BLASTP program in NCBI and KOME against the Arabidopsis key cell cycle proteins (Menges et al. 2005), with the E-value cutoff set as 1e-005. The sequence alignments were then carefully performed with the Vector NTI Advance 9 program.
Phylogenetic analyses were performed through the conserved regions of the alignments. The deduced amino acid sequences were aligned using the computer program Vector NTI Advance 9, and the dendrograms were then generated using the neighbor-joining method.
RT-PCR analysis
Plant tissues of rice were ground in liquid nitrogen. For reverse transcriptase-mediated PCR analysis, total RNA was isolated with the TriPure isolation reagent (Roche Diagnostics) according to the manufacturer’s instructions. The RNA preparation was then treated with DNase I. First-strand synthesis of cDNA was performed by using oligo (dT) primer and M-MLV RT (Promega Corporation). PCR products were fractionated on 1% agarose gels containing ethidium bromide and photographed under UV light. These experiments were independently replicated at least three times under identical conditions. Details of primers are listed in the Supplementary Table 1.
In situ hybridization
In situ hybridization was performed as described by Wang et al. (2006). The plant tissues of rice were fixed in FAA (10% formaldehyde: 5% acetic acid: 50% alcohol) overnight at 4°C. After dehydration, they were embedded in paraplast (Sigma) and sectioned at 8 μm thick. Gene-specific antisense probes of Orysa;CycA2;1(1,351–1,642 bp), Orysa;CycB2;2(1,442-1,712 bp), Orysa;CycU4;4(591–896 bp) and Orysa;CDKG;1(2,512–2,888 bp) were synthesized from PCR products flanked by T7 and Sp6 promoters. They were then hydrolyzed to 200 nucleotides of average length by alkali treatment. After pretreatment, slides were hybridized with 200 ng/ml probes at 42°C overnight in hybridization solution containing 50% formamide. For the detection of hybridized signal, hybridized probes were used with an anti-digoxigenin antibody conjugated with alkaline phosphatase (DIG Nucleic Acid Detection Kit, Boehringer Mannheim). Photographs were taken using the Olympus BH-2 microscope.
Results
Identification of core cell cycle genes in the rice genome
To identify core cell cycle genes in the rice genome, we conducted Blast searches against the rice protein database with query sequences of all previously published core cell cycle regulators from Arabidopsis and a cutoff E-value of 1e-005 (Vandepoele et al. 2002; Wang et al. 2004; Menges et al. 2005). Total of 90 putative core cell cycle proteins were identified through the searches, and they belong to cyclin, CDK, E2F, CKI, CKS, Rb and Wee families, respectively (Supplemental Tables 1 and 2). Of them, 41 were new or corrections of the existing annotations.
Cyclin family
Cyclins are the major factors that determine the timing of CDK activation, and there are 10 types of cyclins in Arabidopsis (A, B, C, D, H, L, T, U, SDS, and J18) (Renaudin et al. 1996; Abrahams et al. 2001; Azumi et al. 2002; Torres Acosta et al. 2004; Wang et al. 2004). In the rice genome, there are 9 different types of cyclins, and the F-type cyclins (CYCF) are unique in rice (La et al. 2006). Except the F-type cyclins, 44 cyclin proteins were identified from the rice genome by using the FGP-MINE programs from the Floral Genome Project Website (http://www.fgp.bio.psu.edu/cgi-bin/fgpmine/fgp_family_list.cgi, Wang et al. 2004). Our results confirmed the previous designation of the cyclins in rice and the evolutionary relationships between the Arabidopsis and rice cyclins (data not shown).
CDK family
CDKs are Ser-Thr protein kinases and involved in the regulation of the eukaryotic cell cycle (King et al. 1994; Lees 1995; Morgan 1995; Pines 1995). To date, 29 Arabidopsis CDK-related sequences were annotated and grouped into 8 different types (Vandepoele et al. 2002; Menges et al. 2005). The most widely conserved CDKs possess a canonical motif in the C-helix, which is involved in the binding the cyclin subunit (De Bondt et al. 1993; Joubès et al. 2000). CDKA has the canonical PSTAIRE motif in the cyclin binding domain (Mironov et al. 1999). B-type CDKs are unique to plants and contain a divergent cyclin binding motif PPTA/TLRE (Fobert et al. 1996; Hirayama et al. 1991; Magyar et al. 1997). In Arabidopsis, CAKs appear to activate CDKs by phosphorylating the threonine residue in the T-loop of the CDKs (Jeffrey et al. 1995; Shimotohno et al. 2003). Three Arabidopsis dimeric CAKs were named CDKD;1–3 and the monomeric CAK was named CDKF;1 (Umeda et al. 1998; Yamaguchi et al. 1998; 2000; Shimotohno et al. 2003). CDKC was characterized by the presence of the PITAIRE motif and CDKE harbours a SPTAIRE motif (Joubès et al. 2000). CDKG was defined by its conserved PLTSLRE motif, and CKL has a (V, I, L)(K, R) FMAREL motif (Menges et al. 2005).
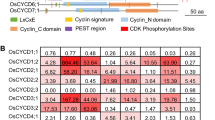
Blast search revealed a total of 25 putative CDK proteins from the rice protein database of NCBI and KOME (Fig. 1A). Among them, 4 CDKs (Orysa;CDKA;1, Orysa;CDKA;2, Orysa;CDKB;2 and Orysa;CDKD;1) have been described previously (Fabian et al. 2000; Hashimoto et al. 1992; Umeda et al. 1999), and 21 were newly described (see Supplemental Table). These CDKs all encompass a catalytic core with short amino- and carboxy-terminal extensions. We further combined the catalytic core with the rest specific motifs in our phylogenetic analysis in order to classify the rice CDK proteins. The results suggested that there were 8 different types of CDKs (A- to G-type and the CKL-type) in the rice genome (Figs. 1A, 2A), similar to those in Arabidopsis (Menges et al. 2005). The number of CDKs in types A, B, D, F, C and CKL was different from that in Arabidopsis (Figs. 1A, 2A). For example, one CDKF was identified in Arabidopsis but 4 rice CDKs (CDKF;1–4) share high homology with the CDKF in Arabidopsis, and 10 CKLs were found in rice compared to 15 CKLs in Arabidopsis (Figs. 1A, 2A). Structural analysis revealed that each type of CDKs contains the similar motifs in Arabidopsis and rice. However, Orysa;CDKC;3 lacks the CDKC specific motif although it shares higher overall sequence similarity with Arath;CDKC;1 (54%) and Arath;CDKC;2 (55%) at the protein level (Fig. 2A).
Phylogenetic tree analysis of CDKs, KRPs and E2Fs in Arabidopsis and rice: (A) Unrooted neighbour-joining tree of all 29 Arabidopsis and 25 rice CDK kinases. Only the protein kinase domains from these proteins were used in this analysis. The proteins referred here were Arath;CKL15, Orysa;CKL4, Arath;CKL7, Arath;CKL8, Arath;CKL9, Orysa;CKL5, Orysa;CKL6, Arath;CKL10, Arath;CKL11, Arath;CKL12, Arath;CKL13, Arath;CKL14, Orysa;CKL9, Orysa;CKL7, Orysa;CKL8, Orysa;CKL10, respectively; (B) unrooted neighbor-joining tree of all 14 KRPs from Arabidopsis and rice after multiple sequence alignment using the full-length protein sequences; (C) unrooted Neighbor-Joining Tree of the E2F, DP and DEL Families, and only the conserved regions with unambiguous alignments were used for phylogenetic analysis
Phylogenetic relationships and structural organization of rice CDKs, KRPs and E2Fs at the protein level: (A) Phylogenetic tree and identification of the conserved functional domains and family-specific protein signatures for rice CDK proteins; (B) phylogenetic tree and the conserved sequence boxes (1–6), PEST motifs and CDK consensus phosphorylation sites of rice KRP proteins were indicated; (C) phylogenetic tree and schematic diagrams showing the various protein domains and boxes as indicated for E2F, DP, and DEL in rice
CKI family
The CKI proteins are the inhibitors of CDKs, and there are two types of CKIs (the INK4 and the Kip/Cip families) in mammals. Only Kip/Cip CDK inhibitors were identified in plants and were designated as Kip-related proteins (KRPs) in Arabidopsis (De Veylder et al. 2001; Vandepoele et al. 2002). The Arabidopsis genome contains 7 KRPs (Arath;KRP1–7), and have been confirmed their inhibitory activity against CDK both in vitro and in vivo (Wang et al. 1997, 1998, 2000; Lui et al. 2000; De Veylder et al. 2001; Zhou et al. 2002; Verkest et al. 2005). Arath;KRP1–7 proteins contain the conserved motifs 1 and 2, including the CDK-binding box, and motif 3 that has the CYCD-binding box (Wang et al. 1998). Recently, Orysa;KRP1–5 have been reported in rice, and Orysa;KRP1 was described in detail (Barrôco et al. 2006).
Here, we identified two other candidates of the Kip/Cip proteins in rice and they were named Orysa;KRP6–7 (Figs. 1B, 2B). The motifs 1, 2 and 3 are located at the C-terminal part of the Orysa;KRP6 protein, whereas, Orysa;KRP7 contains the motifs 4, 5 and 6 at its N-terminal part. Thus, Orysa;KRP7 lacks the CDK-binding box and the CYCD-binding box, suggesting that this gene may be a pseudogene. We further used PEST find software (http://www.emb1.bcc.univie.ac.at/embnet/tools/bio/) to detect the PEST domains which serve as proteolytic signals in KRPs (Rogers et al. 1986). The results indicated that Orysa;KRP6–7 did not contain the PEST domain. Based the phylogenetic analysis of the rice KRPs, Orysa;KRP6 was much closely related to type 1 (Orysa;KRP1–3), whereas Orysa;KRP7 was clustered in type 2 (Orysa;KRP4–5) (Figs. 1B, 2B; Barrôco et al. 2006).
E2F/DP and Rb
Retinoblastoma (Rb/E2F/DP) pathway is conserved among the animal and plant, and controls the G1/S transition. Rb proteins inhibit the activity of E2F factors and have a negative effect on cell cycle progression (Gutierrez et al. 2002). The Rb proteins contain a “pocket” region required for their interactions with other cellular proteins, and the pocket region has two functional domains, RB-A and RB-B, with an intervening spacer (Grafi et al. 1996). A single gene for an Rb-related protein exists in Arabidopsis genome, and is located on chromosome 3. In maize, there are three genes (RBR1, RBR2 and RBR3) as well as differentially spliced transcripts, although their functions are unknown (Grafi et al. 1996; Ach et al. 1997; Hsieh and Wolniak 1998; Sabelli et al. 2005; Sabelli and Larkins 2006). Two homologs of Rb genes (Orysa;Rb1 and Orysa;Rb2) were identified in the rice genome. Both Orysa;Rb1 and Orysa;Rb2 contained RB-A and RB-B domains as verified through protein sequence and structural analysis.
E2F family of transcription factors play important roles in regulating gene expression at the G1/S boundary of the cell division cycle, and acts through a heterodimer with other related proteins such as DP (Castellano et al. 2001; Combettes et al. 1999; De Jager et al. 2001; Vandepeople et al. 2002; Menges et al. 2005). Three E2F, two DP, and three E2F-LIKE (E2F) and DP-LIKE (DEL) genes were identified in the Arabidopsis genome (Magyar et al. 2000; De Jager et al. 2001; Gutierrez et al. 2002; Kosugi and Ohashi 2002). E2Fa,b,c proteins contain a DNA binding domain, a Leucine zipper dimerization domain, a “marked box”, and a Rb binding domain. The two DP proteins have both DNA-binding and Leucine zipper dimerization domains, Whereas three DELs contain only two DNA-binding domains (Vandepeople et al. 2002; Ramirez-Parra et al. 2004). Our studies identified four E2Fs, three DPs, and two DELs in the rice genome by BLAST searches and sequence alignments against the Arabidopsis databases (Figs. 1C, 2C). We found that Orysa;E2F2 and Orysa;E2F4 lacked the Leucine zipper dimerization domain, and Orysa;DP1–3 contained only one DNA-binding domain (Fig. 2C).
CKS and Wee
CDK subunit (CKS) proteins were proposed to act as docking factors that can influence the interactions between the kinase complex and their substrates (Morris et al. 2003). One putative Orysa;CKS was found in the rice genome (Supplementary Table), and the putative protein had a well-conserved N-terminus that shares an overall 95% and 93% similarity with Arabidopsis CKS1 and CKS2, respectively.
The protein kinase Wee inhibits CDKs by phosphorylating a tyrosine residue in the CDK active site (Gould and Nurse 1989; Jin et al. 1996; Sun et al. 1999). One Wee gene (AtWEE1) has been reported in Arabidopsis (Sorrell et al. 2002). In the rice genome, two Wee proteins, Orysa;Wee1 and Orysa;Wee2, were identified, and the two proteins display significant sequence identity with AtWEE1 (50% and 61%, respectively) (Supplementary Table). Like AtWEE1, Orysa;Wee1 has all the sequence characteristics of the Wee kinases: an ATP binding region, including a tyrosine kinase phosphorylation site (Y293) and a Ser/Thr protein kinase active site. A putative nuclear localization signal was found at residue K158 of Orysa;Wee1, whereas Orysa;Wee2 only contains the tyrosine kinase phosphorylation site (Y134).
Expression profiles of rice core cell cycle genes
To understand the functions of rice core cell cycle genes, we investigated their expression patterns in rice tissues by semi-quantitative RT-PCR (Fig. 3). The tissues include roots, young leaves, spikelets, seeds (0, 1, 3, 6 days after pollination or DAP), endosperms (9, 12 and 15 DAP), and embryos (9 and 15 DAP). As shown in Fig. 3A, RNA transcripts were detected in 28 out of 44 cyclin genes. The A- and B-Type cyclin genes were mainly expressed in young seeds and embryos except for Orysa;CycA3;1 and Orysa;CycB1;2. Most D-type cyclin transcripts were accumulated in leaves, spikelets, young seeds, and embryos, although the signals detected were variable in these tissues. Four cyclin genes (Orysa;CycD2;1, Orysa;CycD4;1, Orysa;CycD5;1 and Orysa;CycD6;1) appear to be constitutively expressed in all tissues examined, whereas the Orysa;CycD7;1 transcripts were detectable only in spike tissues. Most of H-, L-, and T-type cyclin genes were likely expressed in all tissues, but the stronger signals were detected in the endosperms and the embryos. Interestingly, Orysa;CycU2;1 and Orysa;CycU3;1 transcripts were mainly detectable only in leaves, indicating a major function of the genes in leaf growth and development.
Expression pattern analysis of the rice core cell cycle genes by RT-PCR in various rice tissues. The rice actin gene was used as an internal control: (A) Cyclin; (B) CDKs and CKS; (C) KRP; (D) E2F/DPs; (E) Rbs. R, root (root tips of seedlings at 1 week after germination); L, young leaf; S, spikelet; 0d–6d, seeds (0, 1, 3, 6 days after pollination, DAP); 9 en–15 en, endosperms (9, 12 and 15 DAP); 9 em and 15 em, embryos (9 and 15 DAP)
The transcripts of 23 CDK genes were detectable by RT-PCR technique. CDKs could be classified into 4 groups according to their expression profiles (Fig. 3B). The largest group is composed of 13 genes (Orysa;CDKA;1, Orysa;CDKA;2, Orysa;CDKB;2, Orysa;CDKC;1, Orysa;CDKC;2, Orysa;CDKC;3, Orysa;CDKD;1, Orysa;CDKE;1, Orysa;CDKF;4, Orysa;CDKG;1, Orysa;CDKG;2, Orysa;CKL6 and Orysa;CKL10), and their transcripts were present in all tissues examined. The second group contains 7 genes (Orysa;CDKB;1, Orysa;CDKF;3, Orysa;CKL2, Orysa;CKL3, Orysa;CKL4, Orysa;CKL5 and Orysa;CKL8), and their transcripts were detected in most of the tissues except the roots. The third group includes two genes: Orysa;CDKF;1 which was expressed in most of tissues except the roots and spikelets, and Orysa;CKL1 whose transcripts were undetectable in the roots and leaves. The last gene, Orysa;CKL7, was exclusively expressed in the ovaries (Fig. 3B).
For E2F/DP and Rb genes, Orysa;E2F1, Orysa;E2F2, Orysa;DP2 and Orysa;DP3 transcripts were not detectable in all tissues collected. In contrast, Orysa;E2F3, Orysa;DEL1, Orysa;Rb1 and Orysa;Rb2 genes were constitutively expressed in all tissues examined. Orysa;E2F4, Orysa;DEL2 and Orysa;DP1 genes were expressed in most of tissues, but not in the roots (Fig. 3C and E). In addition, the expression of three Orysa;CKI genes, such as Orysa;KRP1, Orysa;KRP4 and Orysa;KRP5, were observed in most of tissues examined (Fig. 3D).
Localization of four core cell cycle genes in plant tissues
Four core cell cycle genes were randomly selected for in situ hybridization analysis. Figure 4A–D shows that the signal of Orysa;CycA2;1 transcripts was detected in the tissues of root meristem, shoot meristem, leaf primordium, and young floret, but the signal was weak in the embryo. Compared with Orysa;CycA2;1, Orysa;CycB2;2 transcripts were very abundant in the embryonic tissues, although the signal in other tissues was similar to that of Orysa;CycA2;1 transcripts (Fig. 4E–H). For Orysa;CycU4;4, a weak signal was detected in the root meristem and embryonic tissues compared with that in other tissues examined (Fig. 4I–L). Orysa;CDKG;1 transcripts were accumulated in all tissues examined (Fig. 4M–P). Thus, the in situ hybridization analysis supports the results of our RT-PCR analysis and suggests that the expression of core cell cycle genes was spatially regulated during rice plant development.
Localization of Orysa;CycA2;1, Orysa;CycB2;2, Orysa;CycU4;4 and Orysa;CDKG;1 transcripts as revealed by in situ hybridization. All micrographs show longitudinal sections of rice tissues: (A–D) Localization of Orysa;CycA2;1 mRNA in the rice tissues; (E–H) localization of Orysa; CycB2;2 mRNA in the rice tissues; (I–L) localization of Orysa;CycU4;4 mRNA in the rice tissues; (M–P) localization of Orysa;CycU4;4 mRNA in the rice tissues. (A, E, I and M) root tip; (B, F, J and N) shoot apex; (C, G, K and O) young floret; (D, H, L and P) embryo at 9 DAP. an, anther; ov, ovary; rc, root cap; rd, radicle; rm, root meristem; sam, shoot apical meristem; yl, young leaf. In B, F, J and N bar = 50 μm, in all others bar = 300 μm
Expression of rice core cell cycle genes in response to cytokinin and auxin
Auxins and cytokinins function in many aspects of plant development, such as cell division and expansion, apical dominance, lateral root development and vascular tissue differentiation. (Binns 1994; Leyser 2002; Friml 2003; Mok and Mok 1994, 2001). Previous studies have showed that the expression of many core cell cycle genes was regulated by auxin, cytokinin, and other hormones. For example, auxin was sufficient to upregulate the expression of CDKA;1 in Arabidopsis (Zhang et al. 1996). The CYCA2;2 expression was also induced by auxin during lateral root initiation and elongation in alfalfa (Roudier et al. 2003). Cytokinins are activators of plant cell division both in vivo and in vitro. Treatment with cytokinin led to the upregulation of CycD3 and Triae;CYCD2;1 in Arabidopsis (Riou-Khamlichi et al. 1999; Wang et al. 2006).
To determine which core cell cycle genes respond to cytokinin and auxin, we submerged roots of 2 week-old rice seedlings in a solution of IAA or 6-BA, and then harvested the seedlings at different time. Following treatments with auxin, the expression of the core cell cycle genes was examined for either upregulation or downregulation. Twenty-five genes were found to be upregulated and four genes (Orysa;CycD5;3, Orysa;CycU3;1, Orysa;CDKA;2, and Orysa;CDKD;1) were found to be downregulated (Fig. 5, Supplemental Figure 1). After cytokinin treatment, the transcript levels of 11 genes were increased, and by contrast, the expression of 26 genes was downregulated. The expression of other core cell cycle genes was not regulated by either auxin or cytokinin treatment (Fig. 6, Supplemental Figure 2).
Expression profiles of the rice core cell cycle genes in seedlings after auxin treatment. The relative transcript levels of these genes were analyzed over a 24 h period after 10 μM IAA treatment. Semi-quantitative RT-PCR was performed with three independent biological replicates. The signals of amplified gene products were calculated with the Gel-Pro analyzer (Media Cybernetics, USA). The difference between them is significant (P < 0.05)
Discussion
In this study, we identified and analyzed a large number of rice core cell cycle regulators by using 99 query sequences previously published in Arabidopsis (Vandepoele et al. 2002; Wang et al. 2004; Menges et al. 2005). A total of 90 core cell cycle regulators in the rice genome were identified and they belong to cyclin, CDK, E2F, CKI, Rb, CKS, and WEE families (Figs. 1, 2). Some of the rice core cell cycle regulators were reported previously (Umeda et al. 1999; De Veylder et al. 2001; La et al. 2006). We described and updated 41 regulators in the rice genome (Supplemental Tables 1, 2). Our results indicate that the rice genome contains less number of the core cell cycle regulators than Arabidopsis does, although the rice genome is much larger than the Arabidopsis one. Phylogenetic analysis indicated that 8 different types of CDKs corresponding to those in Arabidopsis identified previously. We also found the corresponding regulators in the rice genome in groups such as E2F, CKI, Rb, CKS, and WEE (Supplemental Tables 1, 2; Figs. 1, 2). The number of regulators varied greatly in most of CDK types and the E2F, CKS, and WEE groups between rice and Arabidopsis. However, the number of the core cell cycle genes is relatively conserved during evolution between the genomes of two species, although the diversification occurred in some types or groups of the core cell cycle genes.
Protein structure analysis showed considerable conservation of the CDK proteins in the catalytic core and the specific motifs for each CDK type except for CDKF in rice (Figs. 1A, 2A). These CDK types were previously identified in Arabidopsis (Vandepoele et al. 2002; Menges et al. 2005). In contrast, KRP proteins were very divergent in their structures (Fig. 2B). For example, Orysa;KRP6 contained only motifs 1, 2, and 3 at its C-terminal part, whereas Orysa;KRP7 had motifs 4, 5, and 6 at its N-terminal part. Although a total of 5 or 6 motifs were identified at both the C- and N-terminal parts of other KRP proteins, only Orysa;KRP4 carries a consensus CDK phosphorylation site (Fig. 2B). The results suggest that Orysa;KRP proteins are less conserved. Among the rice E2F/DP and Rb proteins, DPs and Rbs are relatively conserved, whereas E2F proteins are less conserved as E2F3 lacks a Rb binding box (Fig. 2C).
Expression analysis could provide important information to understand the functions of core cell cycle genes and their molecular action in cell cycle regulation (Wang et al. 2004; Menges et al. 2002, 2003, 2005). According to the expression profiles of the rice core cell cycle genes examined in this study, we can divide them into 5 different classes (Figs. 3, 4). The first class includes 30 genes which were expressed in all tissues. In the second class, the expression of 18 genes was detectable in most of tissues. The third class composed of 10 genes whose transcripts were mainly accumulated in several tissues. The fourth class has 4 genes and their transcripts were specifically accumulated in only one tissue. Finally, the expression of 28 genes was not detected in any tissues for reasons we currently do not understand. Previous study showed that 4 rice CDK genes, cdc2Os1, cdc2Os2, cdc2Os3 and R2 were expressed in roots (Umeda et al. 1999). Transcripts of cdc2Os1, cdc2Os2, and R2 were detected uniformly in the dividing region of the root apex. By contrast, the transcripts of cdc2Os3 were distributed only in patches of the dividing region. These CDK genes in this paper were designated as Orysa;CDKA;1, Orysa;CDKA;2, Orysa;CDKB;2, and Orysa;CDKD;1, respectively, and their transcripts were also detected in roots by RT-PCR (Fig. 3). In addition, the signals of three cyclin-dependent kinase inhibitors, Orysa;KRP1, Orysa;KRP4 and Orysa;KRP5 were accumulated in seeds (Fig. 3), and their expression patterns are in good agreement with those in previous analysis (Barrôco et al. 2006). In Arabidopsis, cyclins were divided into four groups according to their expression profiles (Wang et al. 2004). Further global analyses revealed that most core cell cycle regulators were expressed across almost all tissues and more than 85% of them showed expression at detectable levels in the cell suspension culture (Menges et al. 2005). Therefore, the majority of core cell cycle genes were spatially regulated, and the expression patterns may play a role in plant development. It is interesting that the most closely related core cell cycle genes in many cases showed very similar patterns of expression (Figs. 3, 4) as in the case for the cyclin gene families in Arabidopsis, suggesting possible functional redundancy between the highly similar members (Wang et al. 2004) and functional conservation of the core cell cycle genes between rice and Arabidopsis.
Cell division was also regulated by several plant hormones, and cytokinins and auxins play very important roles in controlling the transcriptional regulation for several core cell cycle genes (Rashotte et al. 2003; Pozo et al. 2005). We analyzed the expression of many core cell cycle genes following treatment with either auxin or cytokinin. The expression of most genes was upregulated by auxin, and the expression of 4 genes was downregulated by auxin (Fig. 5). In contrast, the expression of 11 genes was enhanced by cytokinin and the expression of other 26 genes was decreased following cytokinin treatment (Fig. 6). Combined results of Figs. 5 and 6, expression of 18 genes was regulated by both auxin and cytokinin. Of them, transcript levels of 12 genes were increased following auxin treatment, and however their transcript levels were decreased after cytokinin treatment. It might be very interesting to further study molecular mechanism of the cell cycle gene expression regulated by auxin or cytokinin. Previous study showed that the rice seedlings grown in the dark had decreased expression of the cyclin genes, cycD, cycH1, cycC, cycA1;1 and cycB2;1 (Lee et al. 2006). However, a decrease was prevented by kinetin treatment, indicating that exogenous cytokinin plays an important role in maintaining homeostasis of cyclin gene expression following rapid changes of photoperiod. In Arabidopsis, either exogenous cytokinin or auxin promoted the expression of core cell cycle genes, such as CYCD3, CYCB1 and CDKA (reviewed by Pozo et al. 2005). Our previous study showed that in the tissues of the Arabidopsis plants expressing a wheat cyclin gene, Triae;CYCD2;1, the calli were promoted in the presence of either auxin or cytokinin (Wang et al. 2006). Thus, it is likely that auxin and/or cytokinin may regulate cell cycle through their influences on the transcription of these core cell cycle genes.
Expression profiles of the rice core cell cycle genes in seedlings after cytokinin treatment. The relative transcript levels of these genes were analyzed over a 240 min period after 5 μM cytokinin treatment. Semi-quantitative RT-PCR was performed with three independent biological replicates. The signals of amplified gene products were calculated with the Gel-Pro analyzer (Media Cybernetics, USA). The difference between them is significant (P < 0.05)
References
Abrahams S, Cavet G, Oakenfull EA, Carmichael JP, Shah ZH, Soni R, Murray JA (2001) A novel and highly divergent Arabidopsis cyclin isolated by complementation in budding yeast. Biochim Biophys Acta 1539:1–6
Ach RA, Durfee T, Miller AB, Taranto P, Hanley-Bowdoin L, Zambryski PC, Gruissem W (1997) RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol Cell Biol 17:5077–5086
Azumi Y, Liu D, Zhao D, Li W, Wang G, Hu Y, Ma H (2002) Homolog interaction during meiotic prophase I in Arabidopsis requires the SOLO DANCERS gene encoding a novel cyclin-like protein. EMBO J 21:3081–3095
Barrôco RM, Peres A, Droual AM, De Veylder L, Nguyen le SL, De Wolf J, Mironov V, Peerbolte R, Beemster GT, Inzé D, Broekaert WF, Frankard V (2006) The cyclin-dependent kinase inhibitor Orysa;KRP1 plays an important role in seed development of rice. Plant Physiol 142:1053–1064
Beemster GTS, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inzé D, Vuylsteke M (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 138:734–743
Binns AN (1994) Cytokinin accumulation and action: biochemical, genetic, and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol 45:173–196
Castellano MM, del Pozo JC, Ramirez-Parra E, Brown S, Gutierrez C (2001) Expression and stability of Arabidopsis CDC6 are associated with endoreplication. Plant Cell 13:2671–2686
Combettes B, Reichheld JP, Chaboute ME, Philipps G, Wen HS, Chaubet-Gigot N (1999) Study of phase-specific gene expression in synchronized tobacco cells. Methods Cell Sci 21:109–121
De Bondt HL, Rosenblatt J, Jancarik J, Jones HD, Morgan DO, Kim SH (1993) Crystal structure of cyclin-dependent kinase 2. Nature 363:595–602
De Jager SM, Menges M, Bauer UM, Murray JA (2001) Arabidopsis E2F1 binds a sequence present in the promotor of S-phase-regulated gene AtCDC6 and is a member of a multigene family with differential activities. Plant Mol Biol 47:555–568
De Veylder L, Beeckman T, Beemster GTS, Krols L, Terras F, Landrieu I, Van Der Schueren E, Maes S, Naudts M, Inzé D (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13:1653–1667
De Veylder L, Joubès J, Inzé D (2003) Plant cell cycle transitions. Curr Opin Plant Biol 6:536–543
Fabian T, Lorbiecke R, Umeda M, Sauter M (2000) The cell cycle genes cycA1;1 and cdc2Os-3 are coordinately regulated by gibberellin in planta. Planta 211: 376–383
Fobert PR, Gaudin V, Lunness P, Coen ES, Doonan JH (1996) Distinct classes of cdc2-related genes are differentially expressed during the cell division cycle in plants. Plant Cell 8:1465–1476
Frank M, Schmülling T (1999) Cytokinin cycles cells. Trends Plant Sci 4:243–244
Friml J (2003) Auxin transport—shaping the plant. Curr Opin Plant Biol 6:7–12
Gould KL, Nurse P (1989) Tyrosine phosphorylation of the fission yeast cdc2 + protein kinase regulates entry into mitosis. Nature 342:39–45
Grafi G, Burnett RJ, Helentjaris T, Larkins BA, DeCaprio JA, Sellers WR, Kaelin WG Jr (1996) A maize cDNA encoding a member of the retinoblastoma protein family: Involvement in endoreduplication. Proc Natl Acad Sci U S A 93:8962–8967
Gutierrez C, Ramirez-Parra E., Castellano MM, del Pozo JC (2002) G1 to S transition: more than a cell cycle engine switch. Curr Opin Plant Biol 5:480–486
Hirayama T, Imajuku Y, Anai T, Matsui M, Oka A (1991) Identification of two cell-cycle-controlling cdc2 gene homologs in Arabidopsis thaliana. Gene 105:159–165
Hsieh WL, Wolniak SM (1998) Isolation and characterization of a funcational A-type cyclin from maize. Plant Mol Biol 37:121–129
Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich NP (1995) Mechanism of cdk activation revealed by the structure of a cyclinA-CDK2 complex. Nature 376:313–320
Jin P, Gu Y, Morgan DO (1996) Role of inhibitory CDC2 phosphorylation in radiation-induced G2 arrest in human cells. J Cell Biol 134:963–970
Joubès J, Chevalier C, Dudits D, Heberle-Bors E, Inzé D, Umeda M, Renaudin JP (2000) CDK-related protein kinases in plants. Plant Mol Biol 43:607–620
King RW, Jackson PK, Kirschner MW (1994) Mitosis in transition. Cell 79:563–571
Kosugi S, Ohashi Y (2002) E2F sites that can interact with E2F proteins cloned from rice are required for meristematic tissue-specific expression of rice and tobacco proliferating cell nuclear antigen promoters. Plant J 29:45–59
La H, Li J, Ji Z, Cheng Y, Li X, Jiang S, Venkatesh PN, Ramachandran S (2006) Genome-wide analysis of cyclin family in rice (Oryza Sativa L.). Mol Genet Genomics 275:374–386
Lees E (1995) Cyclin dependent kinase regulation. Curr Opin Cell Biol 7:773–780
Lee H, Auh C-K, Kim D, Lee T-K, Lee S (2006) Exogenous cytokinin treatment maintains cyclin homeostasis in rice seedlings that show changes of cyclin expression when the photoperiod is rapidly changed. Plant Physiol Biochem 44:248–252
Leyser O (2002) Molecular genetics of auxin signaling. Annu Rev Plant Biol 53:377–398
Liu W, Xu ZH, Luo D, Xue HW (2003) Roles of OsCKI1, a rice casein kinase I, in root development and plant hormone sensitivity. Plant J 36:189–202
Lui H, Wang H, Delong C, Fowke LC, Crosby WL, Fobert PR (2000) The Arabidopsis Cdc2a-interacting protein ICK2 is structurally related to ICK1 and is a potent inhibitor of cyclin-dependent kinase activity in vitro. Plant J 21:379–385
Magyar Z, Atanassova A, De Veylder L, Rombauts S, Inzé D (2000) Characterization of two distinct DP-related genes from Arabidopsis thaliana. FEBS Lett 486:79–87
Magyar Z, Meszaros T, Miskolczi P, Deak M, Feher A, Brown S, Kondorosi E, Athanasiadis A, Pongor S, Bilgin M, Bako L, Koncz C, Dudits D (1997) Cell cycle phase specificity of putative cyclin-dependent kinase variants in synchronized alfalfa cells. Plant Cell 9:223–235
Meijer M, Murray JAH (2001) Cell cycle controls and the development of plant form. Curr Opin Plant Biol 4:44–49
Menges M, de Jager SM, Gruissem W, Murray JA (2005) Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J 41:546–566
Menges M, Hennig L, Gruissem W, Murray JA (2002) Cell cycle-regulated gene expression in Arabidopsis. J Biol Chem 277:41987–42002
Menges M, Hennig L, Gruissem W, Murray JA (2003) Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol Biol 53:423–442
Mironov V, De Veylder L, Van Montagu M, Inzé D (1999) Cyclin-dependent kinases and cell division in higher plants—the nexus. Plant Cell 11:509–521
Mironov V, Van Montagu M, Inzé D (1997) Regulation of cell division in plants: an Arabidopsis perspective. Prog Cell Cycle Res 3:29–41
Mok DW, Mok MC (2001) Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol 52:89–118
Mok DWS, Mok MC (1994) Cytokinins: chemistry, activity and function. CRC Press, Boca Raton, FL
Morgan DO (1995) Principles of CDK regulation. Nature 374:131–134
Morris MC, Kaiser P, Rudyak S, Baskerville C, Watson MH, Reed SI (2003) Cks1-dependent proteasome recruitment and activation of CDC20 transcription in budding yeast. Nature 423:1009–1013
Hashimoto J, Hirabayashi T, Hayano Y, Hata S, Ohashi Y, Suzuka I, Utsugi T, Toh-e A, Kikuchi Y (1992) Isolation and characterization of cDNA clones encoding cdc2 homologues from Oryza sativa: a functional homologue and cognate variants. Mol Gen Genet 233:10–16
Pines J (1995) Cyclins and cyclin-dependent kinases: a biochemical view. Biochem J 308:697–711
Potuschak T, Doerner P (2001) Cell cycle controls: genome-wide analysis in Arabidopsis. Curr Opin Plant Biol 4:501–506
Pozo MJ, Van Loon LC, Pieterse CMJ (2005) Jasmonates—signals in plant–microbe interactions. J Plant Growth Regul 23:211–222
Ramirez-Parra E, Lopez-Matas MA, Frundt C, Gutierrez C (2004) Role of an atypical E2F transcription factor in the control of Arabidopsis cell growth and differentiation. Plant Cell 16:2350–2363
Rashotte AM, Carson SDB, To JPC, Kieber JJ (2003) Expression profiling of cytokinin action in Arabidopsis. Plant Physiol 132:1998–2011
Renaudin JP, Doonan JH, Freeman D, Hashimoto J, Hirt H, Inzé D, Jacobs T, Kouchi H, Rouze P, Sauter M, Savoure A, Sorrell DA, Sundaresan V, Murray JA (1996) Plant cyclins: a unified nomenclature for plant A-, B- and D-type cyclins based on sequence organization. Plant Mol Biol 32:1003–1018
Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JAH (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283:1541–1544
Rogers S, Wells R, Rechsteiner M (1986) Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234:364–368
Roudier F, Fedorova E, Lebris M, Lecomte P, Gyorgyey J, Vaubert D, Horvath G, Abad P, Kondorosi A, Kondorosi E (2003) The Medicago species A2-type cyclin is auxin regulated and involved in meristem formation but dispensable for endoreduplication-associated developmental programs. Plant Physiol 131:1091–1103
Sabelli PA, Larkins BA (2006) Grasses like mammals? Redundancy and compensatory regulation within the retinoblastoma protein family. Cell Cycle 5:352–355
Sabelli PA, Dante RA, Leiva-Neto JT, Jung R, Gordon-Kamm WJ, Larkins BA (2005) RBR3, a member of the retinoblastoma-related family from maize, is regulated by the RBR1/E2F pathway. Proc Natl Acad Sci U S A 102:13005–13012
Shimotohno A, Matsubayashi S, Yamaguchi M, Uchimiya H, Umeda M (2003) Differential phosphorylation activities of CDK-activating kinases in Arabidopsis thaliana. FEBS Lett 534:69–74
Sorrell DA, Marchbank A, McMahon K, Dickinson JR, Rogers HJ, Francis D (2002) A WEE1 homologue from Arabidopsis thaliana. Planta 215:518–522
Stals H, Inzé D (2001) When plant cells decide to divide. Trends Plant Sci 6:359–364
Sun Y, Dilkes BP, Zhang C, Dante RA, Carneiro NP, Lowe KS, Jung R, Gordon-Kamm WJ, Larkins BA (1999) Characterization of maize (Zea mays L.) Wee1 and its activity in developing endosperm. Proc Natl Acad Sci U S A 96:4180–4185
Torres Acosta JA, De Almeida Engler J, Raes J, Magyar Z, De Groodt R, Inzé D, De Veylder L (2004) Molecular characterization of Arabidopsis PHO80-like proteins, a novel class of CDKA;1-interacting cyclins. Cell Mol Life Sci 61:1485–1497
Umeda M, Bhalerao RP, Schell J, Uchimiya H, Koncz C (1998) A distinct cyclin-dependent kinase-activating kinase of Arabidopsis thaliana. Proc Natl Acad Sci U S A 95:5021–5026
Umeda M, Umeda-Hara C, Yamaguchi M, Hashimoto J, Uchimiya H (1999) Differential expression of genes for cyclin-dependent protein kinases in rice plants. Plant Physiol 119:31–40
Vandepoele K, Raes J, De Veylder L, Rouze P, Rombauts S, Inzé D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14:903–916
Verkest A., Manes CL, Vercruysse S, Maes S, Van Der Schueren E, Beeckman T, Genschik P, Kuiper M, Inzé D, De Veylder L (2005) The Cyclin-Dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17:1723–1736
Wang F, Huo SN, Guo J, Zhang XS (2006) Wheat D-Type cyclin Triae;CYCD2;1 regulate development of transgenic Arabidopsis plants. Planta 224:1129–1140
Wang G, Kong H, Sun Y, Zhang X, Zhang W, Altman N, dePamphilis CW, Ma H (2004) Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol 135:1084–1099
Wang H, Fowke LC, Crosby WL (1997) A plant cyclin-dependent kinase inhibitor gene. Nature 386:451–452
Wang H, Qi Q, Schorr P, Cutler AJ, Crosby WL, Fowke LC (1998) ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J 15:501–510
Wang H, Zhou Y, Gilmer S, Whitwill S, Fowke LC (2000) Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J 24:613–623
Yamaguchi M, Umeda M, Uchimiya H (1998) A rice homolog of Cdk7/MO15 phosphorylates both cyclin-dependent protein kinases and the carboxy-terminal domain of RNA polymerase II. Plant J 16:613–619
Yamaguchi M, Fabian T, Sauter M, Bhalerao RP, Schrader J, Sandberg G, Umeda M, Uchimiya H (2000) Activation of CDK-activating kinase is dependent on interaction with H-type cyclins in plants. Plant J 24:11–20
Zhang K, Letham DS, John PCL (1996) Cytokinin controls the cell cycle at mitosis by stimulating the tyrosine dephosphorylation and activation of p34cdc2-like H1 histone kinase. Planta 200:2–12
Zhou Y, Fowke LC, Wang H (2002) Plant CDK inhibitors: studies of interactions with cell cycle regulators in the yeast two-hybrid system and functional comparisons in transgenic Arabidopsis plants. Plant Cell Rep 20:967–975
Acknowledgements
This research was supported by the Major State Basic Research Program of the People’s Republic of China (2005CB120803).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material
Fig. S1
Expression profiles of the rice core cell cycle genes in seedlings after auxin treatment. The transcript levels of these genes were analyzed over a 24-h period after 10 μM IAA treatment. The rice actin gene was used as an internal control. (A) Cyclins; (B) CDKs and CKS; (C) KRPs; (D) E2F/DPs; (E) Rbs. ↑, upregulation; ↓, downregulation (JPG 615 kb)
Fig. S2
Expression profiles of the rice core cell cycle genes in seedlings after cytokinin treatment. The transcript levels of these genes were analyzed over 4-h period in response to 5 μM exogenous 6-BA. The rice actin gene was used as an internal control. (A) Cyclins; (B) CDKs and CKS; (C) KRP; (D) E2F/DPs; (E) Rbs. ↑, upregulation; ↓, downregulation (JPG 724 kb)
Table S1
A list of rice core cell cycle genes (DOC 227 kb)
Table S2
Sequences of rice core cell cycle genes (XLS 235 kb)
Rights and permissions
About this article
Cite this article
Guo, J., Song, J., Wang, F. et al. Genome-wide identification and expression analysis of rice cell cycle genes. Plant Mol Biol 64, 349–360 (2007). https://doi.org/10.1007/s11103-007-9154-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-007-9154-y