Abstract
Cell cycle is precisely controlled by numerous regulators. Although extensive studies have identified the cell cycle regulators in rice and Arabidopsis and classified into different gene families, relatively few cell cycle regulators were identified in maize. In this study, 110 putative core cell cycle–related genes in the maize genome were identified through sequence alignment analysis. Among them, 54 members of cyclin family were divided into the A-, B-, C-, D-, F-, L-, T-, and SDS-type subfamilies; 16 members of CDK family were partitioned into the A-, B-, C-, E-, F-, and G-type subfamilies; 19 members of E2F-DP family were separated into the E2F, PAP, DPB, CK, and DEL subfamilies; the remaining genes were nine ICK/KRPs, three EL2/SIMs, three CKSs, five Rbs, and one Wee1 kinase. Maize cell cycle–related genes unequally distributed on all chromosomes, and thirty-six pairs of them were identified as paralogs. The prediction of protein interaction found that 91 proteins were able to interact with each other forming 939 pairs of interactions with a probability more than 0.7. The expression analyses revealed that most of the cell cycle–related genes were expressed in all tissues. The transcription level of some genes in young tissues was higher than that in mature tissues of maize. Meanwhile, some genes from different families shared the similar expression pattern. Overall, the sequence characteristics and expression patterns of maize cell cycle–related genes vary with different gene families. Our results provide basic information for elucidating the functions of these genes in maize.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cell cycle progression is regulated by precise and complex mechanisms that depend on various proteins such as cyclin-dependent kinases (CDKs), cyclins, CDK inhibitors (CKIs) or Kip-related proteins (KRP), cyclin-dependent kinase subunits (CKS), retinoblastoma protein (Rb), E2F/DP transcription factors (E2F/DP), and Wee1 kinase protein (Wee1) (Harry et al. 2006; Piel and Tran 2009). Plant cell cycle is controlled by hormonal, nutritional, and environmental signals (Hartig and Beck 2006; Komaki and Sugimoto 2012; Pozo et al. 2005), and regulatory factors are also highly conserved (De Veylder et al. 2003; Inagaki and Umeda 2011; Polyn et al. 2014).
CDKs are the core regulatory of the cell cycle and have serine/threonine-specific catalytic centers, but their activity depends on cyclins (Inagaki and Umeda 2011; Malumbres 2014). CDKs and cyclins form protein complexes that control various cell cycle phases (Chee and Haase 2010; Cui et al. 2007). CKIs or KRP were first isolated in Arabidopsis by yeast two-hybrid assay and recognized as cyclin-dependent kinase inhibitors (Wang et al. 1997). CKS proteins are docking factors that affect the interactions between the kinase complex and its substrates (Dewitte and Murray 2003). Retinoblastoma-related protein (RBR) regulates cell cycle progression and cellular differentiation (Weinberg 1995). The E2F/DP transcription factor is conserved in plants and shares a conserved DNA-binding domain, which can bind to overlapping sets of target promoters and influence numerous biological processes (Müller and Helin 2000; Van Den Heuvel and Dyson 2008). Wee1 is able to phosphorylate tyrosine residues in CDKs and delay mitosis (Parker and Piwnica-Worms 1992; Russell and Nurse 1987). The aforementioned factors collaboratively regulate the plant cell cycle.
In recent years, the primary cell cycle genes in Arabidopsis and rice had been identified and grouped into the cyclin, CDK, E2F/DP, CKI, Rb, CKS, and WEE1 families (Guo et al. 2007; Menges et al. 2005). Of these, CDKs and cyclins are the most abundant. The A-, B-, C-, D-, H-, L-, T-, U-, SDS-, and J18 cyclin subtypes were found in Arabidopsis (Menges et al. 2005). However, only nine subgroups were detected in rice, including a special F-type (La et al. 2006). The CDK family was divided into seven conservative CDKA-G subfamilies in Arabidopsis and rice, and CDK-like served as a novel subclass, whose function had not been further studied (Guo et al. 2007; Menges et al. 2005; Vandepoele et al. 2002). Prior research revealed that CKIs or KRP, CKS, Rbs, E2F/DPs, and Wee1 existed in the Arabidopsis and rice genomes (Guo et al. 2007; Menges et al. 2005). Besides Arabidopsis and rice, some cell cycle–related genes had been also identified in potato (Yong et al. 2016), tomato (Guo et al. 2010), tobacco (Yu et al. 2003), alfalfa (Horváth 2010), and maize (Godinez-Palma et al. 2013), and they were reported to control cell division and tissue growth and development. Expression pattern analyses of the cell cycle genes disclosed spatiotemporal specificity at the transcriptional level (Beemster et al. 2005; Guo et al. 2007; Menges et al. 2005). Moreover, their expression was induced or repressed by auxins or cytokinins (Guo et al. 2007; La et al. 2006).
Maize is a major global food crop. Its cell cycle regulatory mechanisms have been investigated. The cell cycle–related genes have also been assigned to various families. A-, B-, and D-type cyclins were identified at different maize developmental phases and were significant in maize cell division and tissue development (Godinez-Palma et al. 2013; Hochholdinger et al. 2004; Hsieh and Wolniak 1998; Lara-Núez et al. 2008; Peres et al. 1999; Renaudin et al. 1994; Sun et al. 1999b). Maize A-type CDKs include ZmCdc2a, Zmcdc2b, and ZmCDKA3. ZmCdc2a and Zmcdc2b are highly homologous, ZmCDKA3 was cloned from maize endosperm, and all three are essential for cell proliferation and plant development (Colasanti et al. 1991; Godinez-Palma et al. 2013). Maize KRP genes were first detected in the endosperm and determined to be important for endoreduplication (Coelho et al. 2005). Another five maize ICK/KRP genes were confirmed based on their sequence alignment, and their preliminary in vitro functional characterizations were carried out (Xiao et al. 2017). The RBR proteins ZmRBR1, ZmRBR2, and ZmRBR3 were detected in maize and regulated the cell cycle by interacting with cyclins and E2F/DPs (Ach et al. 1997; Grafi et al. 1996; Sabelli et al. 2005). One Wee1 gene was cloned in a previous study, which could inhibit CDK activity and affected Schizosaccharomyces pombe cell division (Sun et al. 1999a). The maize CKS coding gene was induced by drought (Zhang et al. 2010). The E2F/DP transcription factors were also proved to regulate the maize cell cycle (Sabelli et al. 2005).
Several researchers have studied the function of the core cell cycle–related genes and screened specific gene families in maize genome (Buendia-Monreal et al. 2011; Hu et al. 2010; Sabelli et al. 2005). However, maize cell cycle–related genes have not been completely identified. Here, the known cell cycle–related protein sequences in Arabidopsis and rice are used to screen candidate sets of cell cycle regulators in the maize genome through sequence alignment analysis (Guo et al. 2007; Menges et al. 2005; Vandepoele et al. 2002; Wang et al. 2004). Analyses of conserved domain, phylogeny, and gene duplication are conducted based on their protein sequences. Furthermore, we analyze the co-expression of genes and the interaction probability of proteins and examine their expression patterns in maize tissues. These results will lay a foundation for further study on the mechanism of cell cycle regulation and the functions of core cell cycle genes in maize.
Materials and Methods
Plant Materials
The maize inbred line 18-599, developed by Maize Research Institute of Sichuan Agricultural University, was grown at the university farm during the summer in 2014 and managed according to local maize production standards. When the silks emerged, artificial self-pollination was performed every afternoon. The roots, stems, and leaves were collected at the initial jointing stage. The pollen and filaments were sampled after the tasseling stage before the filaments emerged from the husks. The pericarp, embryo, and endosperm were excised from the seeds 15 days after pollination (DAP). All samples were collected in the afternoon, immediately frozen in liquid nitrogen, and stored at − 70 °C for further use.
Identification of Putative Cell Cycle–Related Genes in the Maize Genome
To identify all putative cell cycle–related proteins in the maize genome, we performed the Hidden Markov Model (HMM) (http://hmmer.janelia.org/) and BLAST searches. The maize proteome sequence (ZmB73_5b_FGS_translations.fasta.gz) was identified from the Maize Genome Sequence Project using the known cell cycle–related protein sequences of rice and Arabidopsis as queries (Guo et al. 2007; Menges et al. 2005). The HMM of the cyclin_N, cyclin_C, Pkinase, and CDI (CDK inhibitor) domains was used to retrieve the maize cyclins, CDK, and ICK/KRP. The E value for the HMMER analysis was set to 1E−10 for maize. The E2F/DPs, Rbs, and other cell cycle genes were identified by blasting the entire maize genome with the known gene sequences of rice and Arabidopsis as queries. All genes retained only one transcript according to a ClustalW 1.83 alignment.
Primary Sequence Analysis and Construction of the Evolutionary Tree in Various Maize Cell Cycle–Related Genes
Multiple sequence analysis was performed by ClustalW 1.83. Evolutionary trees were constructed in MEGA v. 5.10. The neighbor-joining (NJ) method was used with 1000 bootstrap value.
Duplication of Maize Core Cell Cycle–Related Genes
The chromosomal locations of cell cycle–related genes and paralog data were obtained from the Plant Ensemble Database. Chromosomal localization and homology were visualized in Circos (Krzywinski and Schein 2009).
Conserved Domain and Motif Analyses of Various Maize Cell Cycle–Related Genes
To identify the conserved motifs in various core cell cycle–related proteins, we merged the sequences by using GeneStudio. Protein sequences of the maize core cell cycle–related genes were analyzed with MEME (http://meme.nbcr.net/meme/cgi-bin/meme.cgi). The minimum and maximum motif widths and the number of various motifs were 6, 50, and 10, respectively. The conserved domain was predicted by SMART (http://smart.embl-heidelberg.de/). The relationships between the motifs and conserved domains in the sequence were determined by alignment with ClustalW 1.83.
Co-expression Analysis of Cell Cycle–Related Genes
Co-expression analysis of the maize cell cycle–related genes was performed using the genome-wide atlas of transcription during maize development. It included 60 distinct tissues representing 11 major organ systems of the inbred line B73 (GEO No. GSE27004) (Sekhon et al. 2011). Expression data for all cell cycle–related genes were compared pairwise and the Pearson correlation coefficients (PCC) were calculated in Microsoft Excel 2007 (Microsoft Corp, Redmond, WA, USA). R ×64 3.4.1 was used to plot the PCC data.
Predictive Analysis of Cell Cycle–Related Protein Interactions
The String Database (https://string-db.org) was employed to predict cell cycle–related protein interactions. The combined score consisted of gene fusion, homology, co-expression, phylogenetic co-occurrence, experimentally determined interaction, annotated database, automated text-mining, and chromosome neighborhood. The selection threshold was 0.7. Interactions among various cell cycle–related genes were demonstrated in Cytoscape v. 3.7.0 (Demchak et al. 2014).
Total RNA Extraction
Maize tissues were pulverized under liquid nitrogen and mixed with 1 mL TRIzol reagent (Invitrogen, Carlsbad, CA, USA) for mRNA isolation. Trichloromethane was used for removing impurities such as protein and phenol, while isopropanol was used for RNA precipitation. RNA integrity was checked by electrophoresis on agarose gels (1.5% w/v) and ethidium bromide staining. RNA concentrations were determined with NanoDrop™ 2000 (Thermo Fisher Scientific, Waltham, MA, USA).
Semi-Quantitative RT-PCR and Statistical Analysis
First-strand cDNA was generated using 2 μg total RNA with a PrimeScript™ RT reagent kit and a gDNA Eraser (TaKaRa, Dalian, China) following the manufacturer’s instructions. The primers were designed with Primer 5 according to the reference genome sequence (B73 RefGen_v3). The semi-quantitative RT-PCR mix consisted of 10 μL Premix Taq™ (TaKaRa, Dalian, China), 8 μL ddH2O, 5 μM each sense and antisense primers, and 1 μL template cDNA. The semi-quantitative RT-PCR conditions were as follows: one step of 94 °C for 4 min, 94 °C for 30 s, 57–60 °C for 30 s (annealing temperature based on gene primer), 72 °C for 30 s, 28–32 cycles (based on differential gene expression), 72 °C for 5 min, and 12 °C for infinity. These experiments were independently replicated three times under identical conditions. In the independent experiment, the template of all detected genes is the same cDNA. The cloned products were determined by sequencing. They were fractionated on agarose gels (1.5% w/v) containing ethidium bromide and photographed under UV. Primer names, sequences, and cycles are listed in Table S3.
Results
Identification of Putative Cell Cycle–Related Genes in Maize
We performed HMMR analysis and BLAST on the protein sequences of cell cycle–related genes in rice and Arabidopsis to identify maize homologs. The conserved domain was analyzed by SMART. A single-gene transcript was preserved to code the conservative domain in a specific gene family. There were 110 putative core cell cycle–related genes. Of these, there are 54 cyclins, 16 CDKs, 19 E2F/DP transcription factors, 12 ICK and EL2s, three CKSs, five Rbs, and one Wee1s. The maize putative cell cycle–related genes were designated based on the phylogeny of rice cell cycle–related genes (Guo et al. 2007).
CDK Family
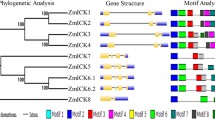
CDKs are the core cell division regulators. Four cell cycle–related genes in the maize genome encoding CDKs (ZmCdc2a, Zmcdc2b, ZmCDKA3, and ZmCDKB1;1) were previously described and were vital to cell proliferation (Colasanti et al. 1991; Godinez-Palma et al. 2013). We identified 16 genes encoding CDKs in the maize genome. We clustered them into the A-, B-, C-, E-, F-, and G-type groups according to their primary sequence alignments and phylogenetic analyses (Fig. 1; Table S1). In maize CDKs, there are three A-type CDKs containing PSTAIRE, two B-type CDKs containing PPTALRE or PPTTLRE, two C-type CDKs containing PITAIRE, one E-type CDK containing SPTAIRE, five F-type CDKs containing SPTAIRE, and three G-type CDKs containing PLTSLRE.
Cyclin Family
Cyclins manage the CDK activation and bind to various CDKs to regulate the cell cycle (Inze and De Veylder 2006). Several A-type, B-type, and D-type cyclins were previously identified in the maize genome (Buendia-Monreal et al. 2011; Godinez-Palma et al. 2013; Hochholdinger et al. 2004; Hsieh and Wolniak 1998; Lara-Núez et al. 2008; Peres et al. 1999; Renaudin et al. 1994; Sun et al. 1999b). Here, we identified 54 genes encoding cyclins and classified them as the A-, B-, C-, D-, F-, L-, T-, and SDS-types based on their phylogenetic analyses (Fig. 1; Table S1) and previous studies on Arabidopsis and rice (La et al. 2006; Menges et al. 2005). The cyclins were distributed on various chromosomes, and different amounts of them occurred in each subgroup.
E2F-DP Transcription Factor
E2F-DP proteins control DNA replication and regulate the cell cycle. They usually transactivate target gene transcription in the form of dimmers (Van Den Heuvel and Dyson 2008; Vandepoele et al. 2002). Nineteen putative genes encoding the E2F domain were recognized in maize and divided into five subgroups for including specific conserved domains (Fig. 1; Table S1). Seven genes were grouped as E2F factors, four as DP factors, and three as DP-E2F-like (DEL) proteins. They contained one E2F, one DP, and two E2F domains, respectively. The remaining five putative genes were categorized into two new groups based on their similarities to DEL or E2F proteins. Three of the five genes were clustered into casein kinase (CK) proteins including a SCOP domain and were highly similar to the DEL protein. Two genes were grouped as purple acid phosphatase (PAP) and were highly analogous to the E2F proteins (Fig. 1; Table S1).
KRP/ICK Protein and SIM/EL2 Proteins
ICK/KRP and SIM/EL2 are important regulators interacting with CDKA (Churchman et al. 2006; De Veylder et al. 2001; Peres et al. 2007; Wang et al. 1997). We identified 12 putative genes encoding ICK/KRP or SIM/EL2 in the maize genome. Nine genes were classified into the ICK/KRP family, which was identified in a previous study (Xiao et al. 2017). Three genes were categorized into the SIM/EL2 subfamily based on their characterizations in rice. ZmEL2 is highly similar to OsEL2. SMR1 and SMR2 closely resemble SIM (Fig. 1; Table S1). ICK/KRP has a conserved CDK inhibitor (CDI) domain at the C-terminal. ZmEL2, SMR1, and SMR2 also have conserved C-terminal sequences but do not form CDK inhibitor (CDI) domains.
RBR/CKS/Wee1
Rbs are involved in many physiological process, such as endoreplication, transcriptional regulation, chromatin remodeling, cell growth, and stem cell differentiation (Desvoyes et al. 2013). Two (Orysa;Rb1 and Orysa;Rb2) and one (Arath;Rb) Rbs have been identified in rice and Arabidopsis, respectively (Guo et al. 2007; Vandepoele et al. 2002). As previous studies have shown, RBR1, RBR2, and RBR3 participate in maize cell cycle regulation (Ach et al. 1997; Grafi et al. 1996; Sabelli et al. 2005). We also recognized two putative genes resembling RBR and designated them as ZmRBRL1 and ZmRBRL2 based on the sequence alignment. At their C-terminals, they were highly similar to maize RBR1/2/3 (Fig. 1; Table S1).
CKS proteins influence the interactions between CDK and their substrates. Two Arabidopsis CKS are associated with cell division and endoreduplication (De Veylder et al. 1997; Vandepoele et al. 2002). The putative rice CKS, Orysa;CKS1, was screened but its function was unknown (Guo et al. 2007). Here, we identified three maize genes that encode CKS proteins (Fig. 1; Table S1).
Protein kinase Wee1 inhibits CDKs by phosphorylating a tyrosine residue in the CDK active site. One Wee1 kinase gene (AtWEE1) in Arabidopsis and two in rice genome were reported (Guo et al. 2007; Sorrell et al. 2002). One maize gene encoding Wee1 protein was identified and found to be important for endosperm development (Sun et al. 1999a). Genome sequencing and prediction showed that the gene encoding the Wee1 protein might consist of two transcripts, Zeama;Wee1 and Zeama;Wee2. The latter lacked a C-terminal sequence (Fig. 1; Table S1).
Primary Sequence Analysis, Evolutionary Tree Construction, and Conserved Motif Analysis for Various Maize Cell Cycle–Related Genes
We used SMART and MEME to search for conserved domains (Fig. 1) and motifs (Fig. 2). The CDKs shared the conserved domain S_TKc, which was essential for protein kinase activity and ATP binding (Fig. 1). Ten conserved motifs in their conserved domains were found in the maize CDKs (Table S2). All cyclins contained the CYCLIN domain and were widely distributed over the C-terminal, N-terminal, and middle regions (Fig. 1). The cyclins also contained the Cyclin_N and BLAST domains. Twenty-eight motifs were discovered in the cyclin domains and their sizes ranged from 10 to 50 amino acids (Table S2). Conserved motifs were distributed mainly in the cyclin domain but also in other positions on the cyclin sequence (Fig. 2).
The E2F_DP domain is a signature sequence localized to the middle region of the maize E2F-DP transcription factors. The E2F and other conserved domains divided the E2F-DP transcription factors into five subgroups (Fig. 1). The E2F group contained a PDB domain near the E2F domain. The DEL group contained two E2F domains. The DPB group contained a DP domain at the C-terminal. The CK subfamily contained SCOP domain, and metallophos domains were specific to the PAP group. MEME search identified 23 conserved motifs in the E2F-DP transcription factor ranging from 15 to 50 amino acids. They were mainly distributed in the conserved domains, but a few were also in the non-conserved domains (Fig. 2).
We found a CDI domain in the maize ICK/KRP gene but no apparent conserved domain in the EL2/SIM group (Fig. 1). A MEME analysis revealed three and five conserved motifs in the ICK/KRP and EL2/SIM gene families, respectively. Among maize ICK/KRP, Zeama;ICK;8 is a special protein with only three motifs compared with others. The remaining eight maize ICK/KRPs share six same conserved motifs (Fig. 2).
The Rbs contained the domains DUF3452, RB_A, CYCLIN, and Rb_C. However, ZmRBRL2 lacked the N-terminal of the DUF3452 domain. RB_A, CYCLIN, and Rb_C are major domains in Rbs, and CYCLIN and Rb_C are located at the C-terminal of protein. The CYCLIN domain of Rb interacts with CDK in cell cycle regulation. Various motifs ranging from 15 to 50 amino acids were identified for the Rb protein domains, which were densely distributed at the C-terminal (Table S2; Fig. 2).
CKS protein contained a widely distributed CKS domain on its sequence. The CKS domains Zeama;CKS;1 and Zeama;CKS;3 were found at the N-terminal, whereas Zeama;CKS;2 was localized to the C-terminal (Fig. 1). Wee1 kinase contained a Pkinase domain on the N-terminal, which is crucial for its function. Zeama;Wee;1 contains the complete domain, while another transcript, Zeama;Wee;2, lacks a part of the Pkinase domain. Meanwhile, the C-terminal Pkinase domain of Wee1 protein is mainly composed of seven conserved motifs (Fig. 2).
The MEME and SMART analyses indicated that the conserved domain contained specific motifs (Fig. 3). According to the MEME analysis of CDKs and Cyclins, motif 1 was a core sequence and constituted the conserved domains S_TKc and CYCLIN. For the ICK/KRP proteins, motifs 1 and 6 comprised the conserved CDI domains. For the E2F-DP transcription factors, the conserved domain E2F contained motif 1 and motif 4. For the remaining gene families, specific motifs also corresponded to particular conserved domains.
Duplications of Maize Core Cell Cycle–Related Genes and Their Mapping on Maize Chromosomes
Genome chromosomal location analyses and primary sequences revealed that maize cell cycle–related genes were unequally distributed over ten chromosomes (Fig. 4). Twenty-four cell cycle–related genes were found on chromosome 1, eight on chromosome 2, seven on chromosome 3, 15 on chromosome 4, 17 on chromosome 5, five on chromosome 6, nine on chromosome 7, 13 on chromosome 9, and six each on chromosomes 8 and 10. Most of the cell cycle–related genes were localized to the ends of the chromosomes. However, the distributions of the gene families on each chromosome varied. Cyclins were widely distributed on all ten chromosomes. The ICK and EL2 were distributed only on chromosomes 1, 4, 5, 8, and 9. Seven chromosomes contain CDK coding genes. Neither chromosome 3 nor 7 bore E2F-DP. CKS, RBR, and Wee1 occurred mainly on chromosomes 1, 2, 3, 4, and 9 (Fig. 4).
We found 36 pairs of paralogs in different families of cell cycle–related genes. They shared a high degree of identity in their protein sequences, which indicated the potential for chromosome duplication. Genes within the same gene family were relatively more likely to undergo duplication. For ICK/KRP and EL2/SIM, all genes except Zeama;EL;2 and Zeama;ICK;8 underwent duplication. For the E2F-DP transcription family, duplication occurred in different subgroups. Duplication also existed among the CDK, cyclin, CKS, and RBR gene families. Genome duplication, unequal chromosomal fragment exchange, and transposable replication might account for these gene duplications. The cell cycle–related gene families were conserved during maize evolution and might be sub-functionalized during cell division.
Co-expression Analysis of Maize Cell Cycle–Related Genes
According to the expression profile in the genome-wide atlas of transcription during maize development, 72 cell cycle–related genes and their expression levels were obtained in various maize tissues. Pearson correlation coefficients between gene pairs were calculated and plotted in Fig. 5. A co-expression clustering analysis grouped cell cycle–related genes into four classes based on their expression levels and correlation coefficients. The latter ranged from − 1 to 1. Zeama;DEL;2 was strongly positively correlated with Zeama;DEL;3 and A-, B-, and D-type cyclins, with the correlation coefficients being around 1. Zeama;DEL;2 was strongly negatively correlated with Zeama;CycT1;1, Zeama;CycT1;2, and Zeama;CDKF;3, with the correlation coefficients being around − 1. Thus, the cell cycle–related genes in maize were co-expressed and their functional interactions and regulatory mechanisms could be predicted.
Predictive Analysis of Maize Cell Cycle–Related Protein Interactions
Ninety-one proteins can interact with each other and form 939 pairs with a probability of more than 0.7 (Table S4; Fig. 6). The A- and B-type CDKs can interact with 63 and 61 proteins, respectively. Zeama;CDKF;1 can interact with 61 proteins. However, Zeama;CDKF;2 can only interact with L-type cyclin. The A-, B-, and SDS-type cyclins can not only interact with themselves but also with A- and B-type CDKs, ICK, E2F, CKS, and Wee1 proteins. Conversely, D-type cyclin can interact mainly with CDKs, RBR, and ICK/KRP proteins. DPB of the E2F-DP transcription factors can interact with E2F and RBR. Nevertheless, E2F can also interact with CDK and A- and F-type cyclins. C-type CDK protein can interact with T-type cyclin. G-type CDK can interact with L-type cyclin. Zeama;ICK;3 and Zeama;ICK;4 can interact with A-type CDK and D-type cyclins (Xiao et al. 2017).
Semi-Quantitative RT-PCR
The functional verification of maize cell cycle–related genes depends on expression analyses. We evaluated the transcription levels of various maize genes by semi-quantitative RT-PCR. The maize tissues included the roots, leaves, stems, developing seeds (10-DAP), anthers, filaments, endosperm (15-DAP), embryos (15-DAP), and pericarps (15-DAP). We measured the expression levels of one Wee1, 13 CDKs, 37 cyclins, 10 ICK/KRP and SIM/ELs, 17 E2F-DP transcription factors, two CKS, and five Rb genes in these tissues. Transcripts of most of these cell cycle–related genes were detected in all tissues but the transcript levels were markedly higher in the younger tissues than in the older tissues (Fig. 7).
Expression pattern analyses by RT-PCR for maize cell cycle–related genes in various tissues. The maize actin gene was used as an internal control. a Cyclin-dependent kinases. b Cyclins. c ICK/KRP and SIM/EL2. d E2F-DPs. e CKS, Rbs, and Wee1. (R) Root. (L) Leaf. (S) Stem harvested at the initial jointing stage. (Se) Seed (10 DAP). (F) Filament. (A) Anther. (P) Pericarp. (Em) Embryo. (En) Endosperm (pericarp, embryo, and endosperm were obtained from 15-DAP seeds)
The 13 CDK genes presented with various expression patterns and were categorized into three groups. The first comprised Zeama;CDKA;1, Zeama;CDKA;2, Zeama;CDKA;3, and Zeama;CDKB;2, which were highly expressed in young maize stems, 10-DAP seeds, and embryos. The second included Zeama;CDKC;1, Zeama;CDKC;2, Zeama;CDKE;1, Zeama;CDKF;1, Zeama;CDKF;3, and Zeama;CDKG;3, which had similar expression levels in all tissues. The third consisted of Zeama;CDKF;5, Zeama;CDKG;1, and Zeama;CDKG;2, which were highly expressed in the anthers (Fig. 7a).
Cyclin expression was very complex. The A-, B-, and especially the D-types had similar and high expression levels in the younger tissues including the stems, 10-DAP seeds, and 15-DAP embryos. Zeama;CycD2;4, Zeama;CycD2;5, and Zeama;CycD2;6 were highly expressed in the anthers. The transcript levels of Zeama;CycD4;5, Zeama;CycD7;1, Zeama;CycL1;1, and T-type cyclin were equal in all tissues. On the other hand, the SDS-type cyclins had relatively low expression levels in all maize tissues (Fig. 7b).
ICK/KRP and SIM/EL2 were expressed in all tissues, but the transcription levels varied. However, they were highly expressed in the young stems, 10-DAP seeds, and embryos (Fig. 7c). Expression data for Zeama;ICK;1 to Zeama;ICK;7 were previously reported (Xiao et al. 2017). The E2F-DP transcription factors had different expression levels in various maize tissues. Zeama;E2F;6 was expressed in the anther and endosperm. Zeama;E2F;7, Zeama;PAP;1, and Zeama;PAP;2 were uniformly expressed in all maize tissues. The other E2F-DP transcription factors were differentially expressed in all tissues but at comparatively higher levels in the younger tissues (Fig. 7d). Zeama;CKS;1 and Zeama;CKS;2 showed similar expression patterns in all tissues (Fig. 7e). Retinoblastoma protein transcripts were detected in all tissues but their levels were comparatively higher in young stems and embryos (Fig. 7e).
Discussion
Plant growth and development are contingent upon cell proliferation. Cell division is precisely controlled by various regulators and influenced by phytohormones and various external signals. Here, we used known Arabidopsis and rice cell cycle–related gene sequences as queries for the identification of cell cycle–related gene in maize (Guo et al. 2007; Menges et al. 2005). We found 110 core cell cycle–related genes in the maize genome, which were classified into the cyclin, CDK, E2F, ICK/KRP, Rb, CKS, and WEE families (Table S1). Certain maize cell cycle–related genes were previously reported including CDKs (Colasanti et al. 1991; Dante et al. 2013), cyclins (Hsieh and Wolniak 1998; Renaudin et al. 1994), ICKs (Coelho et al. 2005), RBR (Ach et al. 1997; Grafi et al. 1996), and Wee1 (Sun et al. 1999a). However, none of these studies comprehensively characterized them in the maize genome.
We revealed that maize had more cell cycle–related genes than rice and Arabidopsis. Nevertheless, all of them belonged to gene families that had been already categorized in the latter two species (Guo et al. 2007; Menges et al. 2005). On the other hand, the distribution of these genes varied with each genome. For example, rice has A-, B-, C-, D-, E-, F-, and G-type CDKs, while maize lacks the D-type CDK (Shimotohno et al. 2004; Takatsuka et al. 2015). The D- and F-type CDKs have the same function as plant CDK-activating kinases (CAK) (Takatsuka et al. 2015). According to primary sequence analysis, maize D-type CDK may be directly classified as an F-type in this study. Various CDKs contain unique conserved motifs that were required for cyclin binding (Mironov et al. 1999). These motifs are also found in maize CDKs, and they are consistent with the results of studies in other plants (Tank and Thaker 2011). A-type CDK is one of the most studied CDK, which is closely related to cell proliferation and tissue development (Hata 1991; Iwakawa et al. 2006; Takashi et al. 1991); B-type CDKs are required for normal cell cycle progression and meristem organization (Andersen et al. 2008); Arabidopsis C-type CDKs can interact with T-type cyclins and play important roles in infection with Cauliflower mosaic virus (CaMV) (Cui et al. 2007). Arabidopsis CDKF;1 participates in activating phosphorylation of cyclin-dependent kinase–activating kinases (Shimotohno et al. 2004). The activity of CDKs depends on cyclins. We classified the 54 maize cyclins into the A-, B-, C-, D-, F-, L-, T-, and SDS-subtypes. In contrast, Arabidopsis has more H-, J18-, and U-type cyclins (Vandepoele et al. 2002). The function of cyclin in different plants is also widely studied. In Arabidopsis, A-type cyclin CYCA2;3 served as a key regulator of ploidy levels (Imai et al. 2006), D-type cyclin CYCD3;1 is the limiting factor for the G1-to-S-phase transition (Menges et al. 2006), and CYCB1 can form complex with CDKB1 to mediate homologous recombination repair (Weimer et al. 2016). Maize cyclins was first cloned in 1994 (Renaudin et al. 1994), A-type cyclins was able to rescue a budding yeast cyclin-deficient mutant (Hsieh and Wolniak 1998), B2-type cyclin is significant in mitotic and endoreduplication (Sabelli et al. 2014), and D-type cyclins are essential for seed germination (Lara-Núez et al. 2008). However, most of the 54 maize cyclins are short of direct functional study (Table S1).
Besides CDK and cyclin, other regulatory factors are also vital to cell cycle. The maize genome comprises 19 E2F-DP genes that are clustered into the E2F, PAP, DPB, CK, and DEL subgroups containing the E2F and other specific conserved domains. It is different from the findings in rice and Arabidopsis, which have nine and eight E2F-DP genes, respectively, and divided into the E2F, DP, and DEL subgroups (Guo et al. 2007; Vandepoele et al. 2002). E2F is mainly associated with Rb by forming a complex to regulate the transcription of genes that are important for differentiation and development (Korenjak and Brehm 2005; Shen 2002). The PAP and CK are the newly discovered E2F proteins in maize, which have their own functional annotations for a specific domain. Thus, they may participate in other biological functions. The CDK inhibitors ICK/KRP and EL2/SIM occur in maize. The eight ICK/KRP genes were equally divided into the B and C subgroups (Torres Acosta et al. 2011; Xiao et al. 2017). Our maize gene classification aligns with that of rice which has seven ICK/KRP genes. Of these, one is identified as a pseudogene (Barroco et al. 2006; Guo et al. 2007; Yang et al. 2011). In contrast, the Arabidopsis ICK/KRP genes are arranged in the A and C subgroups according to the evolutionary analyses of several different species (Torres Acosta et al. 2011). One EL2 and two SIM genes were recognized in maize. Nevertheless, EL2 and SIM occur separately only in rice and Arabidopsis (Churchman et al. 2006; Peres et al. 2007). A sequence alignment analysis of maize EL2/SIM and ICK/KRP showed that they have a conserved C-terminal sequence, but it was not on the same conserved domain. The Rb, CKS, and Wee1 proteins regulate the plant cell cycle. They are found in maize and have been extensively studied in various plant species. Rb proteins participate in the Rb/E2F/DP pathway and negatively affect cell cycle progression (Gutierrez et al. 2002; Shen 2002). Maize RBR1 and RBR2 are able to interact with a plant D-type cyclin (Ach et al. 1997), while RBR1 is regulated by the RBR1/E2F pathway (Sabelli et al. 2005). We identified two new maize Rbs, ZmRBRL1 and ZmRBRL2, and ZmRBRL2 lost N-terminus. The function of both requires further investigation. Arabidopsis CKS binds to the A-type CDKs and participates in mitosis and endoreduplication (De Veylder et al. 1997; Jacqmard et al. 1999). Nevertheless, the specific functions of maize CKS remain unknown. Three CKS genes were identified in the maize genome and they differed in terms of distribution on the CKS domain. Zeama;CKS;1 and Zeama;CKS;1 have similar expression patterns. Wee1 kinase is very essential in cell cycle regulation (Russell and Nurse 1987); it can modulate cell cycle through different mechanism (Tang et al. 1993). In plant, maize Wee1 was first discovered to regulate cell division and CDK kinase activity in the endosperm (Sun et al. 1999a). Arabidopsis Wee1 was reported to arrest yeast cells and make them grow without division. (Sorrell et al. 2002). Whereas, tomato Wee1 could directly control the size of fruit cells (Gonzalez et al. 2007).
Gene expression and co-expression analyses play important roles in the study of gene function. Gene co-expression analysis had been widely used to determine transcriptional regulation in Arabidopsis and rice (Fu and Xue 2010; Persson et al. 2005). Maize genes are also co-expressed in particular physiological pathways (Sekhon et al. 2011; Thirunavukkarasu et al. 2013). Here, we analyzed the correlations among cell cycle–related genes based on existing expression data. We established that co-expression occurs among maize cell cycle–related genes (Fig. 5). Zeama;DEL;2, Zeama;DEL;3, ZmCDKB;2, and A-, B-, and D-type cyclins are strongly positively correlated in co-expression and strongly negatively correlated with Zeama;CycT1;1, Zeama;CycT1;2, and Zeama;CDKF;3. These correlations may help elucidate their functions in the same regulatory pathways. E2F acts on different proteins during cell cycle regulation (Lincker et al. 2008; Veylder et al. 2002). Arabidopsis E2F1 directly binds to the AtCDC6 promoter and regulates G1-to-S progression (Jager et al. 2001); E2F, Rb, and CKI participate in cell cycle signaling in plant-induced immunity (Wang et al. 2014). Gene expression data (Fig. 7) demonstrated that most cell cycle–related gene transcripts occurred in all tissues but at higher levels in the younger than in the older ones. The gene expression and co-expression data and previous results suggested that transcriptional cell cycle regulation was an important direction for future research (Jager et al. 2001). Interactions among regulators were empirically validated and found to regulate the plant cell cycle (Inagaki and Umeda 2011; Inze and De Veylder 2006; Polyn et al. 2014). A combination of the data for protein interaction and co-expression analyses revealed 939 pairs of interaction among 91 proteins with probability of more than 0.7. Some protein interactions have been confirmed. These include the association of D-type cyclin with A-type CDK in maize germination (Godinez-Palma et al. 2013; Lara-Núez et al. 2008) and the interactions among Zeama;ICK;3, Zeama;ICK;4, and A-type CDK in the inhibition of maize CDKA activity (Xiao et al. 2017). The prediction of other protein interactions depends on the results of current investigations of other species. Therefore, another important research focus is the ongoing identification of protein interactions that regulate the cell cycle in maize.
In summary, we present a comprehensive identification and analysis of the maize cell cycle–related genes. Although the gene identification and expression pattern analysis are only parts of the functional progression study, experience in other research has shown that both of them play a fundamental role in cell cycle regulation. These results provide the insight for further study of maize cell cycle regulation.
Abbreviations
- CDK:
-
Cyclin-dependent kinase
- CDKA:
-
A-type CDK
- CDKB:
-
B-type CDK
- CKI:
-
CDK inhibitors
- ICK:
-
Inhibitor of cyclin-dependent kinase
- KRP:
-
Kip-related protein
- EL2/SIM:
-
EL2/SIAMESE
- Rb:
-
Retinoblastoma protein
- E2F/DP:
-
E2F/DP transcription factors
- CKS:
-
Cyclin-dependent kinases subunit
- CDI:
-
CDK inhibitors
- CAK:
-
CDK-activating kinases
- DAP:
-
Day after pollination
- NJ:
-
Neighbor-joining
- PCC:
-
Pearson correlation coefficient
References
Ach RA, Durfee T, Miller AB, Taranto P, Hanley-Bowdoin L, Zambryski PC, Gruissem W (1997) RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol Cell Biol 17:5077–5086
Andersen SU, Buechel S, Zhao Z, Ljung K, Novák O, Busch W, Schuster C, Lohmann JU (2008) Requirement of B2-type cyclin-dependent kinases for meristem integrity in Arabidopsis thaliana. Plant Cell 20:88–100
Barroco RM, Peres A, Droual AM, De Veylder L, Nguyen le SL, De Wolf J, Mironov V, Peerbolte R, Beemster GT, Inze D, Broekaert WF, Frankard V (2006) The cyclin-dependent kinase inhibitor Orysa;KRP1 plays an important role in seed development of rice. Plant Physiol 142:1053–1064
Beemster GT, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inzé D, Vuylsteke M (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 138:734–743
Buendia-Monreal M, Renteria-Canett I, Guerrero-Andrade O, Bravo-Alberto CE, Martinez-Castilla LP, Garcia E, Vazquez-Ramos JM (2011) The family of maize D-type cyclins: genomic organization, phylogeny and expression patterns. Physiol Plant 143:297–308
Chee MK, Haase SB (2010) B-cyclin/CDKs regulate mitotic spindle assembly by phosphorylating kinesins-5 in budding yeast. PLoS Genet 6:e1000935
Churchman ML, Brown ML, Kato N, Kirik V, Hülskamp M, Inzé D, De VL, Walker JD, Zheng Z, Oppenheimer DG (2006) SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell 18:3145–3157
Coelho CM, Dante RA, Sabelli PA, Sun Y, Dilkes BP, Gordon-Kamm WJ, Larkins BA (2005) Cyclin-dependent kinase inhibitors in maize endosperm and their potential role in endoreduplication. Plant Physiol 138:2323–2336
Colasanti J, Tyers M, Sundaresan V (1991) Isolation and characterization of cDNA clones encoding a functional p34cdc2 homologue from Zea mays. Proc Natl Acad Sci U S A 88:3377–3381
Cui X, Fan B, Scholz J, Chen Z (2007) Roles of Arabidopsis cyclin-dependent kinase C complexes in cauliflower mosaic virus infection, plant growth, and development. Plant Cell 19:1388–1402
Dante RA, Sabelli PA, Nguyen HN, Leiva-Neto JT, Tao Y, Lowe KS, Hoerster GJ, Gordon-Kamm WJ, Jung R, Larkins BA (2013) Cyclin-dependent kinase complexes in developing maize endosperm: evidence for differential expression and functional specialization. Planta 239:493–509
De Veylder L, Segers G, Glab N, Casteels P, Van Montagu M, Inzé D (1997) The Arabidopsis Cks1At protein binds the cyclin-dependent kinases Cdc2aAt and Cdc2bAt. FEBS Lett 412:446–452
De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inze D (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13:1653–1668
De Veylder L, Joubes J, Inze D (2003) Plant cell cycle transitions. Curr Opin Plant Biol 6:536–543
Demchak B, Hull T, Reich M, Liefeld T, Smoot M, Ideker T, Mesirov JP (2014) Cytoscape: the network visualization tool for GenomeSpace workflows. F1000Res 3:151
Desvoyes B, de Mendoza A, Ruiz-Trillo I, Gutierrez C (2013) Novel roles of plant retinoblastoma-related (RBR) protein in cell proliferation and asymmetric cell division. J Exp Bot 65:2657–2666
Dewitte W, Murray JA (2003) The plant cell cycle. Annu Rev Plant Biol 54:235–264
Fu FF, Xue HW (2010) Coexpression analysis identifies rice starch regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol 154:927–938
Godinez-Palma SK, Garcia E, Sanchez Mde L, Rosas F, Vazquez-Ramos JM (2013) Complexes of D-type cyclins with CDKs during maize germination. J Exp Bot 64:5661–5671
Gonzalez N, Gévaudant F, Hernould M, Chevalier C, Mouras A (2007) The cell cycle-associated protein kinase WEE1 regulates cell size in relation to endoreduplication in developing tomato fruit. Plant J 51:642–655
Grafi G, Burnett RJ, Helentjaris T, Larkins BA, DeCaprio JA, Sellers WR, Kaelin WG (1996) A maize cDNA encoding a member of the retinoblastoma protein family: involvement in endoreduplication. Proc Natl Acad Sci U S A 93:8962–8967
Guo J, Song J, Wang F, Zhang XS (2007) Genome-wide identification and expression analysis of rice cell cycle genes. Plant Mol Biol 64:349–360
Guo J, Kwon HK, Wang MH (2010) Characterization of three A-type cyclin genes in tomato (Solanum lycopersicum ) treated with auxins. Appl Biol Chem 53:266–274
Gutierrez C, Ramirez-Parra E, Castellano MM, del Pozo JC (2002) G 1 to S transition: more than a cell cycle engine switch. Curr Opin Plant Biol 5:480–486
Harry E, Monahan L, Thompson L (2006) Bacterial cell division: the mechanism and its precision. Int Rev Cytol 253:27–94
Hartig K, Beck E (2006) Crosstalk between auxin, cytokinins, and sugars in the plant cell cycle. Plant Biol 8:389–396
Hata S (1991) cDNA cloning of a novel cdc2+/CDC28-related protein kinase from rice. FEBS Lett 279:149–152
Hochholdinger F, Woll K, Sauer M, Dembinsky D (2004) Genetic dissection of root formation in maize (Zea mays) reveals root-type specific developmental programmes. Ann Bot 93:359–368
Horváth GV (2010) Activation of an alfalfa cyclin-dependent kinase inhibitor by calmodulin-like domain protein kinase. Plant J 46:111–123
Hsieh W-L, Wolniak SM (1998) Isolation and characterization of a functional A-type cyclin from maize. Plant Mol Biol 37:121–129
Hu X, Cheng X, Jiang H, Zhu S, Cheng B, Xiang Y (2010) Genome-wide analysis of cyclins in maize (Zea mays). Genet Mol Res 9:1490–1503
Imai KK, Ohashi Y, Tsuge T, Yoshizumi T, Matsui M, Oka A, Aoyama T (2006) The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell 18:382–396
Inagaki S, Umeda M (2011) Cell-cycle control and plant development. Int Rev Cell Mol Biol 291:227–261
Inze D, De Veylder L (2006) Cell cycle regulation in plant development. Annu Rev Genet 40:77–105
Iwakawa H, Shinmyo A, Sekine M (2006) Arabidopsis CDKA; 1, a cdc2 homologue, controls proliferation of generative cells in male gametogenesis. Plant J 45:819–831
Jacqmard A, Veylder LD, Segers G, Engler JA, Bernier G, Montagu MV, Inze D (1999) Expression of CKS1At in Arabidopsis thaliana indicates a role for the protein in both the mitotic and the endoreduplication cycle. Planta 207:496–504
Jager SMD, Menges M, Bauer UM, Murray JAH (2001) Arabidopsis E2F1 binds a sequence present in the promoter of S-phase-regulated gene AtCDC6 and is a member of a multigene family with differential activities. Plant Mol Biol 47:555–568
Komaki S, Sugimoto K (2012) Control of the plant cell cycle by developmental and environmental cues. Plant Cell Physiol 53:953–964
Korenjak M, Brehm A (2005) E2F-Rb complexes regulating transcription of genes important for differentiation and development. Curr Opin Genet Dev 15:520–527
Krzywinski M, Schein JI (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645
La H, Li J, Ji Z, Cheng Y, Li X, Jiang S, Venkatesh PN, Ramachandran S (2006) Genome-wide analysis of cyclin family in rice (Oryza sativa L.). Mol Gen Genomics 275:374–386
Lara-Núez A, Nd J, Vázquez-Ramos JM (2008) Maize D4;1 and D5 cyclin proteins in germinating maize. Associated kinase activity and regulation by phytohormones. Physiol Plant 132:79–88
Lincker F, Roa H, Lang J, Sanchez-Calderon L, Smetana O, Cognat V, Keller M, Mediouni C, Houlné G, and Chabouté ME (2008) Plant E2F factors in cell cycle, development and DNA damage response.17-31
Malumbres M (2014) Cyclin-dependent kinases. Genome Biol 15:122
Menges M, De Jager SM, Gruissem W, Murray JA (2005) Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J 41:546–566
Menges M, Samland AK, Planchais S, Murray JA (2006) The D-type cyclin CYCD3;1 is limiting for the G1-to-S-phase transition in Arabidopsis. Plant Cell 18:893–906
Mironov V, Veylder LD, Montagu MV, Inzé D (1999) Cyclin-dependent kinases and cell division in plants: the nexus. Plant Cell 11:509–521
Müller H, Helin K (2000) The E2F transcription factors: key regulators of cell proliferation. Biochim Biophys Acta (BBA) Rev Cancer 1470:M1–M12
Parker LL, Piwnica-Worms H (1992) Inactivation of the p34cdc2-cyclin B complex by the human WEE1 tyrosine kinase. Science 257:1955–1957
Peres A, Ayaydin F, Nikovics K, Gutiérrez C, Horváth GV, Dudits D, Fehér A (1999) Partial synchronization of cell division in cultured maize (Zea mays L.) cells: differential cyclin, cdc2, histone, and retinoblastoma transcript accumulation during the cell cycle. J Exp Bot 50:1373–1379
Peres A, Churchman ML, Hariharan S, Himanen K, Verkest A, Vandepoele K, Magyar Z, Hatzfeld Y, Van Der Schueren E, Beemster GT, Frankard V, Larkin JC, Inze D, De Veylder L (2007) Novel plant-specific cyclin-dependent kinase inhibitors induced by biotic and abiotic stresses. J Biol Chem 282:25588–25596
Persson S, Wei H, Milne J, Page GP, Somerville CR (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci U S A 102:8633–8638
Piel M, Tran PT (2009) Cell shape and cell division in fission yeast. Curr Biol 19:823–827
Polyn S, Willems A, De Veylder L (2014) Cell cycle entry, maintenance, and exit during plant development. Curr Opin Plant Biol 23:1–7
Pozo JCD, Matas L, Parra R, Gutierrez C (2005) Hormonal control of the plant cell cycle. Physiol Plant 123:173–183
Renaudin J-P, Colasanti J, Rime H, Yuan Z, Sundaresan V (1994) Cloning of four cyclins from maize indicates that higher plants have three structurally distinct groups of mitotic cyclins. Proc Natl Acad Sci U S A 91:7375–7379
Russell P, Nurse P (1987) Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell 49:559–567
Sabelli PA, Dante RA, Leiva-Neto JT, Jung R, Gordon-Kamm WJ, Larkins BA (2005) RBR3, a member of the retinoblastoma-related family from maize, is regulated by the RBR1/E2F pathway. Proc Natl Acad Sci U S A 102:13005–13012
Sabelli PA, Dante RA, Nguyen HN, Gordon-Kamm WJ, Larkins BA (2014) Expression, regulation and activity of a B2-type cyclin in mitotic and endoreduplicating maize endosperm. Front Plant Sci 5:561
Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM (2011) Genome-wide atlas of transcription during maize development. Plant J 66:553–563
Shen WH (2002) The plant E2F-Rb pathway and epigenetic control. Trends Plant Sci 7:505–511
Shimotohno A, Umeda-Hara C, Bisova K, Uchimiya H, Umeda M (2004) The plant-specific kinase CDKF;1 is involved in activating phosphorylation of cyclin-dependent kinase-activating kinases in Arabidopsis. Plant Cell 16:2954–2966
Sorrell DA, Marchbank A, McMahon K, Dickinson RJ, Rogers HJ, Francis D (2002) A WEE1 homologue from Arabidopsis thaliana. Planta 215:518–522
Sun Y, Dilkes BP, Zhang C, Dante RA, Carneiro NP, Lowe KS, Jung R, Gordon-Kamm WJ, Larkins BA (1999a) Characterization of maize (Zea mays L.) Wee1 and its activity in developing endosperm. Proc Natl Acad Sci U S A 96:4180–4185
Sun Y, Flannigan BA, Setter TL (1999b) Regulation of endoreduplication in maize (Zea mays L.) endosperm. Isolation of a novel B1-type cyclin and its quantitative analysis. Plant Mol Biol 41:245–258
Takashi H, Yoshiro I, Toyoaki A, Minami M, Atsuhiro O (1991) Identification of two cell-cycle-controlling cdc2 gene homologs in Arabidopsis thaliana. Gene 105:159–165
Takatsuka H, Umeda-Hara C, Umeda M (2015) Cyclin-dependent kinase-activating kinases CDKD;1 and CDKD;3 are essential for preserving mitotic activity in Arabidopsis thaliana. Plant J 82:1004–1017
Tang Z, Coleman TR, Dunphy WG (1993) Two distinct mechanisms for negative regulation of the Wee1 protein kinase. EMBO J 12:3427–3436
Tank JG, Thaker VS (2011) Cyclin dependent kinases and their role in regulation of plant cell cycle. Biol Plant 55:201–212
Thirunavukkarasu N, Hossain F, Mohan S, Shiriga K, Mittal S, Sharma R, Singh RK, Gupta HS (2013) Genome-wide expression of transcriptomes and their co-expression pattern in subtropical maize (Zea mays L.) under waterlogging stress. PLoS One 8:e70433
Torres Acosta JA, Fowke LC, Wang H (2011) Analyses of phylogeny, evolution, conserved sequences and genome-wide expression of the ICK/KRP family of plant CDK inhibitors. Ann Bot 107:1141–1157
Van Den Heuvel S, Dyson NJ (2008) Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol 9:713–724
Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14:903–916
Veylder LD, Beeckman T, Beemster GTS, Engler JDA, Ormenese S, Maes S, Naudts M, Schueren EVD, Jacqmard A, Engler G (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa–DPa transcription factor. EMBO J 21:1360–1368
Wang H, Fowke LC, Crosby WL (1997) A plant cyclin-dependent kinase inhibitor gene. Nature 386:451–452
Wang G, Kong H, Sun Y, Zhang X, Zhang W, Altman N, Ma H (2004) Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol 135:1084–1099
Wang S, Gu Y, Zebell SG, Anderson LK, Wang W, Mohan R, Dong X (2014) A noncanonical role for the CKI-RB-E2F cell-cycle signaling pathway in plant effector-triggered immunity. Cell Host Microbe 16:787–794
Weimer AK, Biedermann S, Harashima H, Roodbarkelari F, Takahashi N, Foreman J, Guan Y, Pochon G, Heese M, Van DD (2016) The plant-specific CDKB1-CYCB1 complex mediates homologous recombination repair in Arabidopsis. EMBO J 35:2068–2086
Weinberg RA (1995) The retinoblastoma protein and cell cycle control. Cell 81:323–330
Xiao Q, Zhang C, Li H, Wei B, Wang Y, Huang H, Li Y, Yu G, Liu H, Zhang J (2017) Identification and functional analysis of the ICK gene family in maize. Sci Rep 7:43818
Yang R, Tang Q, Wang H, Zhang X, Pan G, Tu J (2011) Analyses of two rice (Oryza sativa) cyclin-dependent kinase inhibitors and effects of transgenic expression of OsiICK6 on plant growth and development. Ann Bot 107:1087–1101
Yong R, Hwang KR, Cho S, Lee M, Kil EJ, Choi S, Hahn BS, Kim D, Auh CK, Lee S (2016) Expression analysis of D-type cyclin in potato (Solanum tuberosum L.) under different culture conditions. Acta Physiol Plant 38-47:36
Yu Y, Steinmetz A, Meyer D, Brown S, Shen WH (2003) The tobacco A-type cyclin, Nicta;CYCA3;2, at the nexus of cell division and differentiation. Plant Cell 15:2763–2777
Zhang ZB, Min LU, Hui-Yong LI, Zhang DF, Liu YH, Shi YS, Song YC, Wang TY, Li Y (2010) Isolation and expression analysis of a drought-induced gene “ZmCKS2” in maize(Zea mays L.). Acta Agron Sin 36:945–952
Acknowledgments
We acknowledge the use of the Dr. Rajandeep S. Sekhon (Department of Energy Great Lakes Bioenergy Research Center, University of Wisconsin-Madison, USA) expression profile to do the co-expression analysis.
Funding
This work was supported by the National Natural Science Foundation of China (No: 31571757) and the National Key Basic Research Program of China (No: 2014CB138202).
Author information
Authors and Affiliations
Contributions
Y.H., Q.X, and B.W. designed the study. Q.X. and Y.W. performed the experiments. B.W. and H.H. analyzed the data. Y.H., Q.X., and B. S. AJAYO prepared and revised the manuscript, and all the authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key Message
• Core cell cycle–related gene identification and expression analysis in maize provided basic information for cell cycle regulation research in maize.
Rights and permissions
About this article
Cite this article
Xiao, Q., Wei, B., Wang, Y. et al. Core Cell Cycle–Related Gene Identification and Expression Analysis in Maize. Plant Mol Biol Rep 39, 72–86 (2021). https://doi.org/10.1007/s11105-020-01236-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-020-01236-9