Abstract
Purpose
This prospective study aimed to evaluate the effects of genetic polymorphisms in sulindac-related metabolizing enzyme genes including FMO3 and AOX1 on the population pharmacokinetics of sulindac in 58 pregnant women with preterm labor.
Methods
Plasma samples were collected at 1.5, 4, and 10 h after first oral administration of sulindac. Plasma concentrations of sulindac and its active metabolite (sulindac sulfide) were determined, and pharmacokinetic analysis was performed with NONMEM 7.3.
Results
The mean maternal and gestational ages at the time of dosing were 32.5 ± 4.4 (range, 20–41) years and 27.4 ± 4.4 (range, 16.4–33.4) weeks, respectively. In the population pharmacokinetic analysis, one depot compartment model of sulindac with absorption lag time best described the data. The metabolism of sulindac and sulindac sulfide was described using Michaelis-Menten kinetics. In stepwise modeling, gestational age impacted volume of distribution (Vc), and FMO3 rs2266782 was shown by the Michaelis constant to affect conversion of sulindac sulfide to sulindac (KM32); these were retained in the final model.
Conclusions
Genetic polymorphisms of FMO3 and AOX1 could affect the pharmacokinetics of sulindac in women who undergo preterm labor. The results of this study could help clinicians develop individualized treatment plans for administering sulindac.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Preterm birth is defined as birth before the completion of 37 weeks of gestational age. Preterm delivery is the principal cause of neonatal mortality and morbidity and has major socioeconomic implications (1). Numerous factors have been associated with preterm labor, but the cause of more than 45% of cases of spontaneous preterm birth remains unknown (2).
Sulindac, a nonsteroidal anti-inflammatory drug that inhibits cyclooxygenase, is used as a tocolytic agent to prolong pregnancy in women at risk of preterm labor. Sulindac is generally administered orally, and 90% of the dose is absorbed via passive diffusion (3). Sulindac is a prodrug that is metabolized into two metabolites, mainly in the liver and kidneys (4,5,6,7). Sulindac with the p-methylsulfinyl group is reversibly converted to the sulfide metabolite, which is an active form of prostaglandin synthesis inhibitor (8). Alternatively, sulindac can be irreversibly oxidized to sulindac sulfone, an inactive form. Subsequently, sulindac and sulindac sulfone are conjugated as an ester glucuronide (9). Although 40%–60% of sulindac, the sulfone metabolites, and their conjugates are eliminated by the kidney, (3) the sulfide form is mainly re-oxidized to sulindac; no significant level of free or conjugated sulfide was detected in human urine samples (10). Also, all three chemical compounds are entero-hepatically recycled, and approximately 25% of an administered dose is eliminated in the feces. The absolute values of biliary clearance for the three compounds have large inter-individual variation; however, the relative ratio is constant, averaging 1:12:12 for sulindac sulfide:sulindac:sulindac sulfone (11).
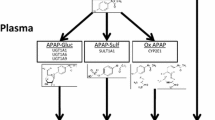
It is known that sulindac undergoes reduction to sulindac sulfide by aldehyde oxidase (AOX) (12). Inversely, flavin-containing monooxygenase 3 (FMO3) oxidizes sulindac sulfide to sulindac, and sulindac to sulindac sulfone, inactivating metabolism of the drug (13) (Fig. 1).
It is well known that genetic polymorphisms lead to inter-individual differences in plasma drug concentrations and enzyme activities. FMO3 enzyme activity displays inter-individual and inter-ethnic differences according to FMO3 gene polymorphisms (14). We previously reported the effects of polymorphisms of FMO3 on the area under the curve (AUC) of sulindac sulfide plasma concentrations using the Winnolin program (15). However, the results required further validation because sampling points were not extensive. Furthermore, despite the potential role of AOX in sulfoxide reduction, (12,16) little is known about the role of AOX in drug metabolism.
Therefore, the present study aimed to establish a population pharmacokinetic model of sulindac and its active metabolite (sulindac sulfide) in pregnant women who experience preterm labor, considering covariates such as FMO3 and AOX1 genetic polymorphisms and the demographic characteristics of the patients.
Materials and Methods
Patients and Data Collection
Patients were recruited at Ewha Womans University Mokdong Hospital between 2010 and 2013. Inclusion criteria were: patient age ≥ 18 years, gestational age between 16 and 37 weeks, uterine contractions with a frequency of 1 per 12 min, and evident cervical changes. Exclusion criteria were: severe pre-eclampsia, fetal/placental/amniotic abnormalities, congenital anomalies, and fetal distress. Because participants were intending to prolong pregnancy, sample collection and plasma analysis were performed depending on the patients’ schedules, and afterward the data were pooled for population analysis. A total of 58 patients were included. All patients gave written informed consent before participating in the study. This study was approved by the Ethics Committee of the Ewha Womans University Mokdong Hospital Institutional Review Board (IRB No. 217–1-26).
All patients received 200 mg sulindac (Imbaron®; Sam Jin Pharm., Seoul, Korea) orally twice a day. Blood and plasma samples were collected at 1.5, 4, and 10 h after the first dose of sulindac. All samples were stored at −70°C until analysis.
Data on the following patient characteristics were collected: maternal age, gestational age at drug administration, body weight, type of gestation, mode of delivery, cervical dilation and effacement at the time of drug administration, fetus presentation, number of neonatal births, and medications used concomitantly with sulindac.
Sulindac and sulindac sulfide concentrations in plasma were analyzed by high-performance liquid chromatography (HPLC) according to our previously reported methods (15). The limits of quantification of sulindac and sulindac sulfide were 0.5 and 0.3 μg/mL, respectively.
Genotyping Analysis
Genomic DNA of the study patients was prepared from EDTA-blood samples using the QIAamp DNA Blood Mini Kit (QIAGEN GmbH, Hilden, Germany). Genetic information about the FMO3 and AOX1 genes was obtained from the International Haplotype Mapping Project website and the Haploview program. There were 23 SNPs in the FMO3 gene and 5 SNPs in the AOX1 gene with minor allele frequencies of ≥10% in Japanese and Chinese populations, respectively. To validate linkage disequilibrium (LD) information, the HaploReg version 3.0 program, which is based on the 1000 Genomes Project, was used (17). Five tagged SNPs in the FMO3 gene (rs909530, rs2266782, rs1800822, rs1736557, and rs2075992) captured the common variations within the gene and the surrounding area with a minimum R2 of 0.80. The rs2266780 SNP (18,19,20) tagged with rs2266782 was also included, based on the results of a previous study (15). Ultimately, a total of six SNPs in the FMO3 gene were included in the study. Further, four SNPs in the AOX1 gene (rs2293528, rs10497853, rs10931910, and rs2241080) were added by HaploReg and NCBI/dbSNP. All told, a total of 10 SNPs were investigated. We used genotyping results for 42 patients from our previous study (15) for four FMO3 SNPs (rs2266780, rs1736557, rs2266782, and rs909530), which were merged with genotyping results of 16 more patients. For all other SNPs, genotyping was done for all 58 patients.
The genotypes of 5 SNPs in the FMO3 gene (rs909530, rs2266780, rs2266782, rs1736557, and rs2075992) were examined by a single base primer extension assay using the ABI PRISM SNaPshot Multiplex kit (Applied Biosystems, Foster City, CA, USA). Five SNPs (FMO3 [rs1800822] and AOX1 [rs2293528, rs10497853, rs10931910, and rs2241080]) were investigated by the TaqMan allelic discrimination technique. RT-PCR was accomplished with an ABI 7300 instrument (Applied Biosystems, Carlsbad, CA, USA).
Pharmacokinetic Analysis
Pharmacokinetic analyses were conducted using the nonlinear, mixed-effect modeling approach implemented in NONMEM version 7.3 (ICON Development Solutions, Hanover, MD, USA). The plasma concentration-time profiles for sulindac and the metabolite were explained using the PREDPP subroutine ADVAN6 (general non-linear model).
One- and two-compartment models with first-order absorption and elimination, with or without absorption lag time, were compared, allowing for various forms of metabolism kinetics. Model selection was guided by the adequacy of parameter estimates, objective function value (OFV), and goodness-of-fit plots. The estimation method used was a first-order conditional estimation method with an interaction option (FOCE+I).
Because the inter-conversion of sulindac to sulindac sulfide is reversible, the best basic model was established simultaneously for sulindac and sulindac sulfide. The inter-individual variability of the pharmacokinetic parameters was estimated by an exponential error model and was expressed as Pi = TVP ∗ exp(ηi) where Pi, the individual parameter for the ith subject, was expressed as a function of the typical value of parameter (TVP). Individual deviation, represented by ηi, is a random pharmacokinetic parameter value that is independent and normally distributed with mean 0 and variance ω2.
The residual errors of the pharmacokinetic parameters were estimated with a proportional error model. SIGMA was one fixed by variance for EPS (1) without scale, and an additional one of THETA was used for residual error proportional scaling (https://www.cognigencorp.com/nonmem/nm/99may232001.html). The residual error is expressed as:
where Cij and \( \mathrm{C}{\prime}_{ij}^2 \) are the jth observed and predicted concentration values in the ith individual, respectively. θ and θ’ are the standard deviation of the proportional residual error and the additional residual error, respectively.
The covariates investigated to explain the inter-patient and residual variability of pharmacokinetic parameters of sulindac and sulindac sulfide in this study were: gestational age at dosing, body weight, and genetic polymorphisms in FMO3 (rs909530, rs2266780, rs2266782, rs1800822, rs1736557, and rs2075992) and AOX1 (rs2293528, rs10497853, rs10931910, and rs2241080). Missing covariates of continuous values were replaced with median values calculated in the pooled dataset. In the case of missing categorical covariates, all of the information was excluded from the analysis dataset. Additionally, omitted plasma concentrations of parent and metabolite were also excluded.
The correlation between covariates and pharmacokinetic parameters was evaluated using the stepwise covariate model (SCM) implemented in PsN. During forward selection, a covariate was selected if it showed a significant decrease under χ2 distribution (p − value = 0.05) and incorporated an increased number of parameters in OFV relative to the structural model when added to the base model. According to the SCM process, the relationship between all PK parameters and covariates were evaluated one-by-one in the first step; we evaluated those against the structural model in this step. The significant relations between covariates and PK parameters were retrieved as the reference model for the second stage, and the other covariates were added to this model one-by-one again for evaluation of additional relations. This process was repeated until no covariate reached the defined statistical significance. Finally, we used the model that included all significant relationships as the full model.
All variables found to have a significant influence on the model were included in the full model simultaneously. Subsequently, each covariate was re-evaluated through backward elimination and the criterion for the backward elimination step was an increase of >6.63 (χ2 test, p − value < 0.01) in OFV. The covariates that resulted in a significant increase in OFV were retained in the final model.
To evaluate the accuracy and robustness of the final model, bootstrap, a resampling method, and visual predictive check validation (VPC, n = 1000) were conducted using PsN (ver. 4.70) and R (ver. 3.5.0). The 95% confidence intervals for the 5th, 50th, and 95th percentiles were obtained from the simulation results. The 5th, median, and 95th percentiles of observed concentrations were plotted against time to be compared with the median and 90% prediction intervals from the final model. The bootstrap procedure was repeated 1000 times from the original dataset. The medians and 5th – 95th percentiles of the bootstrap result were compared with the predicted population parameter values from the original dataset.
Statistical Analysis
All statistical analyses were conducted with the Statistical Package for Social Sciences Version 20.0 for Windows (SPSS 20.0 K, SPSS INC., Chicago, IL, USA). The Hardy-Weinberg Equilibrium (HWE) test was performed using the Chi-square test. A p − value < 0.05 was considered statistically significant.
Results
The characteristics of 58 patients included in the present study are listed in Table 1. The mean maternal age was 32.5 ± 4.4 years (range: 20–41 years). Among the patients, 38 (65.5%) were over 26 weeks of gestational age at the time of dosing, and mean body weight was 60.2 ± 9.7 kg (range: 45.0–92.0 kg). The numbers of fetuses that were presented as vertex, breech, and transverse were 36, 11, and 3 respectively. Five patients had twin pregnancies and one patient had triplets.
Seventeen patients had normal delivery, and 23 delivered babies by cesarean section. Ritodrine and magnesium sulfate were administered concomitantly with sulindac to 35 (60.3%) and 7 (12.1%) patients among the 58 patients, respectively. Twenty (34.5%) patients had full-term deliveries, while 22 (37.9%) had preterm deliveries (before 37 weeks).
Six single nucleotide polymorphisms (SNPs) in FMO3 and 4 SNPs in AOX1 were analyzed. The allelic frequencies of the studied genes and the HWE are shown in Supplementary table. All the observed frequencies were consistent with HWE.
A total of 156 sulindac and 165 sulindac sulfide concentrations were measured. Among them, 22 concentrations (6.8%) were excluded because they were below limit of quantitation. Consequently, 146 sulindac concentrations and 153 sulindac sulfide concentrations were included in the population pharmacokinetic modeling. After screening the one- and two-compartment pharmacokinetic models, the one-compartment pharmacokinetic model for sulindac and sulindac sulfide with first-order absorption and elimination was selected. For the absorption model, the one depot compartment model for sulindac with lag time was the optimal fit for our data. According to Michaelis-Menten kinetics, the metabolism model of reversible biotransformation of sulindac and sulindac sulfide was described optimally in the collected data. The central compartment volume of distribution for sulindac sulfide was assumed to be equal to that of the parent central compartment because there was little difference between the two structures. The schematic basic model describing the pharmacokinetics of sulindac and its active metabolite is represented in Fig. 2.
Schematic basic model of sulindac CL/F: population mean value of clearance, Vparent/F: volume of distribution of sulindac, ka: absorption rate constant, k20: elimination rate constant of sulindac, Km23: Michaelis constant of the conversion of sulindac to sulindac sulfide, Km32: Michaelis constant of the conversion of sulindac sulfide to sulindac, Tlag: lag time, Vmax23: maximum reaction velocity of the conversion of sulindac to sulindac sulfide, Vmax32: maximum reaction velocity of the conversion of sulindac sulfide to sulindac.
Demographic and genetic polymorphism data were assessed through the SCM building procedure. Each SNP was categorized into 2 groups: wild homozygote/heterozygote and variant homozygote. In the stepwise forward selection, the following covariates that showed a significant decrease (p − value < 0.05) in the OFV value were included: AOX1 rs10497853 and GA26 (gestational age divided by 26 weeks) for Vc/F, AOX1 rs2241080 for KM23, and FMO3 rs2266780 and rs2266782 for KM32. In the backward elimination step, only GA26 for Vc/F and FMO3 rs2266782 for KM32 were retained under the criteria of p − value < 0.01. No covariates significantly influenced CL/F and KM23.
The descriptive parameters in the final model are summarized in Table 2. The absorption rate constant of sulindac (ka), the volume of distribution of the central compartment of sulindac and its metabolite (Vc/F), and clearance (CL/F) were 0.352 h−1, 14.8 L, and 17.1 L/h, respectively. The maximum reaction velocity of the conversion from sulindac to sulindac sulfide (Vmax23) and the maximum reaction velocity of the conversion from sulindac sulfide to sulindac (Vmax32) were 1390 mg/h and 6.27 mg/h, respectively. KM23 (Michaelis constant of the conversion of sulindac to sulindac sulfide) and KM32 (Michaelis constant of the conversion of sulindac sulfide to sulindac) were 1640 mg/L and 9.46 mg/L, respectively. The absorption lag time of sulindac (Tlag) in the gastrointestinal tract was 0.498 h.
Diagnostic plots of the final model, including OBS (observed concentration) vs. PRED (population predictions), OBS vs. IPRED (individual predictions), CWRES (conditional weighted residuals) vs. PRED, and CWRES vs. time are shown in Fig. 3. The diagnostic plots indicate acceptable goodness of fit. The CWRES for the predicted concentrations were distributed symmetrically and were mostly within 3 units of the null ordinate, presenting a good fit of the model to the original dataset.
Diagnostic plots of the final model for sulindac and sulindac sulfide (a) OBS (observed concentration) vs. PRED (population predictions) (b) OBS (observed concentration) vs. IPRED (individual predicted concentration) (c) CWRES (conditional weighted residuals) vs. PRED (population predictions) (d) CWRES (conditional weighted residuals) vs. time Dotted line, line of unity.
The inter-individual variabilities in Vc/F, CL/F, KM23, and KM32 were 270.8%, 61.3%, 42.8%, and 30.5%, respectively, respectively, as shown in Table 2. The shrinkage values for Vc/F, CL/F, KM23, and KM32 were 19.1%, 14.3%, 22.3%, and 19.9%, respectively.
To evaluate the final model, visual predictive check validation (VPC, n = 1000) simulations and 1000 bootstrap analyses were conducted. For VPC, most data on sulindac and sulindac sulfide fitted well within the 5th to 95th percentiles. As shown in Fig. 4, the final model was more accurate and precise than the base model in describing the pharmacokinetics of the drug, especially for sulindac sulfide. Among the 1000 bootstrap analyses, 57.3% ran successfully; the 5th and 95th percentile results are presented in Table2.
Visual predictive check (VPC) of the final population pharmacokinetic models (a) VPC of sulindac, (b) VPC of sulindac sulfide. Observation data (circles), blue solid lines, 5th and 95th percentiles of observed concentrations; red solid lines, 50th percentile of observed concentrations; semitransparent field, 95% confidence intervals of the simulated 5th, 50th, 95th percentiles from 1000 simulations.
Pharmacokinetic simulations were performed for 4 scenarios, considering two significant covariates in the final model to assess clinical practicality. We created four datasets of 1000 patients, setting fixed and random model effects as equal to those of the final model. Next, AUC0 → ∞ and AUC0 → 24hr values of sulindac sulfide were calculated for each simulated result. As shown in Table 3, the estimated AUC0 → ∞ and AUC0 → 24hr were not affected by the covariate of GA26. In contrast, genotypes of FMO3 rs2266782 had a large difference in estimated AUC0 → ∞ and AUC0 → 24hr. Simulations which included the FMO3 rs2266782 AA type population showed an approximately five-fold increase in AUC0 → ∞ and AUC0 → 24hr of sulindac sulfide.
Discussion
In this study, a population pharmacokinetic model of sulindac and its active metabolite in pregnant Korean women was developed. Covariates that might influence pharmacokinetic parameters were also identified. The one-compartment pharmacokinetic model adequately explained the pharmacokinetics of sulindac and its metabolite in pregnant women undergoing preterm labor. Also, the impact of gestational age on Vc/F and FMO3 polymorphisms on KM32 was comprehensively assessed.
A sulindac population pharmacokinetic model has been previously developed in healthy subjects (21). In that study, the population pharmacokinetics were characterized by a seven-compartment model featuring enterohepatic recirculation. Several useful estimates in healthy subjects were suggested, and demographic factors such as sex and body weight were evaluated to explain inter-individual variability. However, the pharmacokinetics were only partially explained, and the estimates cannot be applied to pregnant women. In terms of the effects of drug-metabolizing enzyme polymorphisms on sulindac pharmacokinetics, a study in healthy Chinese volunteers reported that variant-type homozygote carriers of both rs2266780 (E308G) and rs2266782 (E158K) showed significantly higher systemic exposure (Cmax and AUC) levels of the drug. (22).
To our knowledge, this is the first study to examine the population pharmacokinetics of sulindac in pregnant women. In prodrugs such as sulindac, the activity of metabolizing enzymes has an important role in drug effects, and large inter-individual variability could result from genetic polymorphisms of drug-metabolizing enzymes. Also, considering the physiological changes during pregnancy, insight into differences in sulindac pharmacokinetics of pregnant women with different drug-metabolizing enzyme genotypes is relevant for clinical practice.
The estimates of CL/F and Vc/F from the final model were 17.1 L/h and 14.8 L, respectively. The CL/F of the parent drug was higher than estimated in another study (3), possibly because of the physiological changes in pregnancy. The glomerular filtration rate (GFR), on which renal excretion of drugs depends, is increased by 50% in the first trimester and continued to increase during pregnancy (23,24). It then decreases during the last 3 weeks of pregnancy and reaches its postpartum value by the last week of pregnancy (23,25). The activity of uridine diphosphate glucuronosyltransferase (UGT) isoenzymes, which are phase 2 enzymes, is increased during pregnancy, (23) suggesting that the CL of sulindac and sulindac sulfone may be increased. Because our study population consisted of pregnant women with preterm labor, the GFR values and the activity of UGT enzymes were likely higher than those of normal, healthy women.
The CL of sulindac sulfide was neglected in our model because the average CL ratio of sulindac to sulfide is approximately 12:1 (3). As there are only a few structural differences between sulindac and sulindac sulfide, we assumed that the volume of distribution of sulindac is equal to that of sulindac sulfide.
The inter-individual variability of Vc/F, expressed as a percentage of the coefficient of variation, was 270.8%, which is consistent with the large variations in volume of distribution observed in pregnant women. Due to increases in water, fat, and protein, women gain approximately 11.3 kg or more during pregnancy (26). Also, starting at 6–8 weeks of gestational age and peaking at 32 weeks, maternal blood volume increases by 40%–50% compared to that of non-pregnant women (27). Because the subjects of the present study had different gestational ages, the inter-individual variability in Vc/F was high. The inter-individual variability of Vc/F in the base model was 332.6%, and it decreased in the final model, which took covariates into account. Although we speculated that physiological changes in pregnant women, such as those involving water, protein, and fat, caused high variability of Vc, no association was found between body weight and Vc or CL. Further research is required to validate our findings.
The covariate modeling results also showed that gestational age has a significant association with Vc/F. Our study participants were categorized into two groups according to gestational age under 26 weeks or 26 weeks and older, as 26 weeks was the mode value in the study population.
In the metabolism component of our final model, the biotransformation between sulindac and sulindac sulfide was described by Michaelis-Menten kinetics. FMO enzymes catalyzing N-oxidation and S-oxidation in human liver microsomes were reported to follow Michaelis-Menten kinetics in previous studies (28,29,30). AOX enzymes in the human liver and skin were also shown to follow Michaelis-Menten kinetics (31,32).
Controversy remains whether to set the volume of the metabolite at the same value as the parent drug. We identified both volumes and selected a statistically significant model. Further, since the metabolite can return to the parent drug form, we judged it more appropriate to set the same value for volume of distribution of the parent and the metabolite.
FMO3 rs2266782 had a significant association with KM32. FMOs belong to a family of flavoprotein enzymes that catalyze the oxidation of endogenous and exogenous compounds, including drugs (12). FMO3 has been identified as the major isoform in the human liver, intervening in the NADPH-dependent oxidative metabolism of numerous heteroatom-containing chemicals, especially those possessing nitrogen, sulfur, phosphorus, or selenium as their site of oxidation (15,33,34). Several variations in the FMO3 gene have been reported, and many studies have illustrated the effects of FMO3 polymorphisms on the metabolism of drugs, including sulindac, ranitidine, methimazole, and benzydamine (35,36,37,38).
Among the FMO3 polymorphisms, the rs2266782 (E158K) mutation reportedly reduces the activity of the enzyme (28,36,37). Also, an in vitro study showed that among several FMO SNPs, rs2266782 (E158K) had the greatest effect on the Kcat value of Michaelis-Menten kinetics. In a 3D model, E158 was found in a putative access channel to the catalytic domain near the NADPH-binding motif. Although capacity of the variants is substrate dependent, E158K variants had lower Km and clearance for clomiphene metabolism than wild-type (39). . Gene mutations leading to amino acid sequence changes may alter the affinity of an enzyme for its substrate. In our study, the KM32 of the homozygous variant (AA type) group was 3.9-fold higher than that of the homozygous wild/heterozygous (GG/GA type) group. An elevated Km of the metabolism step may have reduced the activity of FMO3, causing a low binding affinity for the substrate, which is in agreement with previously reported results (40).
Despite the positive value of impact of FMO3 rs2266782 on K32 in the final RSEmodel, because the histogram of impact of K32-FMO3 rs2266782 from the bootstrap showed an additional peak, zero was included (red box, Supplementary figure). Because our data were collected from 58 pregnant women and also showed large variance among them, we speculate that the additional peak may be simply due to a specific subject with a larger variance, when resampling proceeded for the bootstrap.
In contrast to FMO3, none of the genotype covariates of AOX1 was found to be significant. AOXs are molybdo-flavoenzymes that exhibit reductase activities on zonisamide, sulindac, and imipramine N-oxide (11,41). Among the several AOX genes, AOX1 is the main isoform in humans (34). A previous study on the reduction of sulindac in rats revealed that the AOXs are responsible for sulfoxide reduction in liver and kidney tissues (15). Although it is known that human AOX is much more active than rodent AOX, (42) genetic variations of AOX1 did not show any significant effects.
Because GA26 was a significant covariate of Vc/F, which does not directly affect AUC0 → ∞ or AUC0 → 24hr, the differences in AUCs between groups with different GA26 types were not very large. In contrast, simulated estimates of the AUC0 → ∞ and AUC0 → 24hr values of the population with FMO3 rs2266782 AA type revealed large increases in both values. As FMO3 rs2266782 had a significant effect on KM32, the AA type population had a larger KM32 value than did the other group (GG/GA type). Populations with high KM32 were found to have reduced FMO3 enzyme activity. As a result, the AUC0 → ∞ and AUC0 → 24hr values of sulindac sulfide in the population with the AA genotype were estimated to be five-fold higher than the values in the population with the GG/GA genotype.
This study has several limitations. First, the sampling schedule was so sparse that the inter-individual variability in the absorption phase could not be taken into account; however, some individual plots showed the absorption phases, from which Tmax could be obtained. Therefore, the absorption rate constant and lag time could be determined from the population mean. According to a previous report by Berg et al., the absorption process of sulindac is considered fast, with high bioavailability, lag time, and entero-hepatic recirculation (EHR) of drug (21). Despite sparse sampling in the absorption phase, each PK estimate was similar to that in the model of Berg et al., and VPC showed good performance of the model. However, as a double peak pattern was not observed in our dataset, EHR was not applied in the final model. Second, sulindac is not the first-line treatment in pregnant women with preterm labor because of its oral dosing. In our institute, sulindac is used as an add-on therapy for patients who are not treated effectively with ritodrine injection. Thus, it was not possible to obtain the pharmacodynamic phenotype of sulindac therapy alone. Third, although the plasma concentrations of sulindac sulfone could yield valuable quantitative information about sulindac metabolism, we did not measure sulfone concentrations.
In summary, the current study is the first to investigate the population pharmacokinetics of sulindac in women with preterm labor, considering both clinical and genetic variation factors as covariates. In our study population, women with homozygous variants of FMO3 rs2266782 (AA type) showed a higher KM32 for the metabolism from sulindac sulfide to sulindac, signifying low enzyme affinity for the substrate. The results of this study can be used to develop individualized treatment plans. By determining FMO3 genotype at the beginning of treatment, clinicians may be able to tailor regimens for effective use of sulindac in patients with preterm labor.
Abbreviations
- AOX :
-
Aldehyde oxidase
- CWRES:
-
Conditional weighted residuals
- FMO3 :
-
Flavin-containing monooxygenase 3
- HWE :
-
Hardy-Weinberg equilibrium
- OFV:
-
Objective function value
- SNP :
-
Single nucleotide polymorphism
- SCM:
-
Stepwise covariate model
- VPC:
-
Visual predictive check
References
Simhan HN, Caritis SN. Prevention of preterm delivery. N Engl J Med. 2007;357:477–87.
Miyazaki C, Moreno GR, Ota E, Swa T, Oladapo OT, Mori R. Tocolysis for inhibiting preterm birth in extremely preterm birth, multiple gestations and in growth-restricted fetuses: a systematic review and meta-analysis. Reprod Health. 2016;13:4.
Davies NM, Watson MS. Clinical pharmacokinetics of sulindac. A dynamic old drug. Clin Pharmacokinet. 1997;32:437–59.
Eriksson LO, Bostrom H. Deactivation of sulindac-sulphide by human renal microsomes. Pharmacol Toxicol. 1988;62:177–83.
Duggan DE, Hooke KF, Hwang SS. Kinetics of the tissue distributions of sulindac and metabolites. Relevance to sites and rates of bioactivation. Drug Metab Dispos. 1980;8:241–6.
Miller MJ, Bednar MM, McGiff JC. Renal metabolism of sulindac: functional implications. J Pharmacol Exp Ther. 1984;231:449–56.
Ratnayake JH, Hanna PE, Anders MW, Duggan DE. Sulfoxide reduction. In vitro reduction of sulindac by rat hepatic cytosolic enzymes. Drug Metab Dispos. 1981;9:85–7.
Brunell D, Sagher D, Kesaraju S, Brot N, Weissbach H. Studies on the metabolism and biological activity of the Epimers of Sulindac. Drug Metab Dispos. 2011;39:1014–21.
Kuehl GE, Lampe JW, Potter JD, Bigler J. Glucuronidation of nonsteroidal anti-inflammatory drugs: identifying the enzymes responsible in human liver microsomes. Drug Metab Dispos. 2005;33:1027–35.
Duggan DE, Hare LE, Ditzler CA, Lei BW, Kwan KC. The disposition of sulindac. Clin Pharmacol Ther. 1977;21:326–35.
Dujovne C, Pitterman A, Vincek W, Dobrinska M. Enerohepatic circulation of sulindac and metabolites. Clin Pharmcol Ther. 1983;33:172–7.
Kitamura S, Ohashi KNK, Sugihara K, Hosokawa R, Akagawa Y, Ohta S. Extremely high drug-reductase activity based on aldehyde oxidase in monkey liver. Biol Pharm Bull. 2001;24:856–9.
Hisamuddin IM, Yang VW. Genetic polymorphisms of human flavin-containing monooxygenase 3: implications for drug metabolism and clinical perspectives. Pharmacogenomics. 2007;8:635–43.
Shimizu M, Yano H, Nagashima S, Murayama N, Zhang J, Cashman JR, et al. Effect of genetic variants of the human flavin-containing monooxygenase 3 on N- and S-oxygenation activities. Drug Metab Dispos. 2007;35:328–30.
Park S, Lee NR, Lee KE, Park JY, Kim YJ, Gwak HS. Effects of single-nucleotide polymorphisms of FMO3 and FMO6 genes on pharmacokinetic characteristics of sulindac sulfide in premature labor. Drug Metab Dispos. 2014;42:40–3.
Lee SC, Renwick AG. Sulphoxide reduction by rat and rabbit tissues in vitro. Biochem Pharmacol. 1995;49:1557–65.
Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9.
Park CS, Kang JH, Chung WG, Yi HG, Pie JE, Park DK, et al. Ethnic differences in allelic frequency of two flavin-containing monooxygenase 3 (FMO3) polymorphisms: linkage and effects on in vivo and in vitro FMO activities. Pharmacogenetics. 2002;12:77–80.
Koukouritaki SB, Poch MT, Henderson MC, Siddens LK, Krueger SK, VanDyke JE, et al. Identification and functional analysis of common human flavin-containing monooxygenase 3 genetic variants. J Pharmacol Exp Ther. 2007;320:266–73.
Lattard V, Zhang J, Tran Q, Furnes B, Schlenk D, Cashman JR. Two new polymorphisms of the FMO3 gene in Caucasian and African-American populations: comparative genetic and functional studies. Drug Metab Dispos. 2003;31:854–60.
Berg AK, Mandrekar SJ, Ziegler KL, Carlson EC, Szabo E, Ames MM, et al. Population pharmacokinetic model for cancer chemoprevention with sulindac in healthy subjects. J Clin Pharmacol. 2013;53:403–12.
Tang Y, Hu K, Huang W, Wang C, Liu Z, Chen Y, et al. Effects of FMO3 polymorphisms on pharmacokinetics of sulindac in Chinese healthy male volunteers. Biomed Res Int. 2017;4189678.
Anderson GD. Pregnancy-induced changes in pharmacokinetics: a mechanistic-based approach. Clin Pharmacokinet. 2005;44:989–1008.
Davison JM, Dunlop W. Renal hemodynamics and tubular function normal human pregnancy. Kidney Int. 1980;18:152–61.
Davison JM, Dunlop W, Ezimokhai M. 24-hour creatinine clearance during the third trimester of normal pregnancy. Br J Obstet Gynaecol. 1980;87:106–9.
Tasnif Y, Morado J, Hebert MF. Pregnancy-related pharmacokinetic changes. Clin Pharmacol Ther. 2016;100:53–62.
Hytten FE, Paintin DB. Increase in plasma volume during normal pregnancy. J Obstet Gynaecol Br Emp. 1963;70:402–7.
Stormer E, Roots I, Brockmoller J. Benzydamine N-oxidation as an index reaction reflecting FMO activity in human liver microsomes and impact of FMO3 polymorphisms on enzyme activity. Br J Clin Pharmacol. 2000;50:553–61.
Wang L, Christopher LJ, Cui D, Li W, Iyer R, Humphreys WG, et al. Identification of the human enzymes involved in the oxidative metabolism of dasatinib: an effective approach for determining metabolite formation kinetics. Drug Metab Dispos. 2008;36:1828–39.
Lickteig AJ, Riley R, Melton RJ, Reitz BA, Fischer HD, Stevens JC. Expression and characterization of functional dog flavin-containing monooxygenase 3. Drug Metab Dispos. 2009;37:1987–90.
Manevski N, Balavenkatraman KK, Bertschi B, Swart P, Walles M, Camenisch G, et al. Aldehyde oxidase activity in fresh human skin. Drug Metab Dispos. 2014;42:2049–57.
Al-Salmy HS. Individual variation in hepatic aldehyde oxidase activity. IUBMB Life. 2001;51:249–53.
Hernandez D, Addou S, Lee D, Orengo C, Shephard EA, Phillips IR. Trimethylaminuria and a human FMO3 mutation database. Hum Mutat. 2003;22:209–13.
Hines RN, Cashman JR, Philpot RM, Williams DE, Ziegler DM. The mammalian flavin-containing monooxygenases: molecular characterization and regulation of expression. Toxicol Appl Pharmacol. 1994;125:1–6.
Mayatepek E, Flock B, Zschocke J. Benzydamine metabolism in vivo is impaired in patients with deficiency of flavin-containing monooxygenase 3. Pharmacogenetics. 2004;14:775–7.
Hisamuddin IM, Wehbi MA, Schmotzer B, Easley KA, Hylind LM, Giardiello FM, et al. Genetic polymorphisms of flavin monooxygenase 3 in sulindac-induced regression of colorectal adenomas in familial adenomatous polyposis. Cancer Epidemiol Biomark Prev. 2005;14:2366–9.
Hisamuddin IM, Wehbi MA, Chao A, Wyre HW, Hylind LM, Giardiello FM, et al. Genetic polymorphisms of human flavin monooxygenase 3 in sulindac-mediated primary chemoprevention of familial adenomatous polyposis. Clin Cancer Res. 2004;10:8357–62.
Kang JH, Chung WG, Lee KH, Park CS, Kang JS, Shin IC, et al. Phenotypes of flavin-containing monooxygenase activity determined by ranitidine N-oxidation are positively correlated with genotypes of linked FM03 gene mutations in a Korean population. Pharmacogenetics. 2000;10:67–78.
Catucci G, Bortolussi S, Rampolla G, Cusumano D, Gilardi G, Sadeghi S. Flavin-containing monooxygenase 3 polymorphic variants significantly affect clearance of tamoxifen and clomiphene.
Fields PA, Houseman DE. Decreases in activation energy and substrate affinity in cold-adapted A4-lactate dehydrogenase: evidence from the Antarctic notothenioid fish Chaenocephalus aceratus. Mol Biol Evol. 2004;21:2246–55.
Garattini E, Terao M. The role of aldehyde oxidase in drug metabolism. Expert Opin Drug Metab Toxicol. 2012;8:487–503.
Barr JT, Choughule K, Jones JP. Enzyme kinetics, inhibition, and regioselectivity of aldehyde oxidase. Methods Mol Biol. 2014;1113:167–86.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) funded by the Korean government (MEST) (grant number: NRF-2010-0022544) and the research fund of Hanyang University (grant number: HY-2018-N).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interests
No potential conflict of interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sung, J.W., Yun, Hy., Park, S. et al. Population Pharmacokinetics of Sulindac and Genetic Polymorphisms of FMO3 and AOX1 in Women with Preterm Labor. Pharm Res 37, 44 (2020). https://doi.org/10.1007/s11095-020-2765-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11095-020-2765-6