Abstract
A fundamental research topic in domain adaptation is how best to evaluate the distribution discrepancy across domains. The maximum mean discrepancy (MMD) is one of the most commonly used statistical distances in this field. However, information about distributions could be lost when adopting non-characteristic kernels by MMD. To address this issue, we devise a new distribution metric named maximum mean and covariance discrepancy (MMCD) by combining MMD and the proposed maximum covariance discrepancy (MCD). MCD probes the second-order statistics in reproducing kernel Hilbert space, which equips MMCD to capture more information compared to MMD alone. To verify the efficacy of MMCD, an unsupervised learning model based on MMCD abbreviated as McDA was proposed and efficiently optimized to resolve the domain adaptation problem. Experiments on image classification conducted on two benchmark datasets show that McDA outperforms other representative domain adaptation methods, which implies the effectiveness of MMCD in domain adaptation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
In standard machine learning, both training and test data are assumed to be drawn from the same distribution. However, this assumption turns out unrealistic for lots of real-world problems, which could cause performance degradation for testing. For example, in visual recognition task, test images may differ from training ones due to changes in backgrounds, sensors, viewpoints, etc. To address this issue, domain adaptation has been proposed in quest of adapting the models built in one domain (source domain) to serve another different but related domain (target domain), in such a way that the learned models perform well in the target domain.
In DA problems, the dataset from the source domain and the dataset from the target domain usually follow different distributions. To narrow the difference, the first thing is to appropriately evaluate the distribution discrepancy across domains for most DA models. Many candidate statistical distances, such as the Kullback–Leibler divergence [5], the Bregman divergence [33], the Wasserstein distance [32] and the maximum mean discrepancy (MMD) [11], can be used to achieve this purpose. MMD is one of the most widely used distances and it is designed to evaluate the distance between the kernel mean embedding of distributions in a reproducing kernel Hilbert space (RKHS). MMD possesses a decent theoretical property, i.e., characteristic kernels establish MMD as metrics on the space of probability distributions [9, 11, 34]. Due to its non-parametric form and its theoretical properties, MMD has attracted widespread attention from the DA community.
Much progress has been made in an effort to explore MMD and thereby obtain transferrable knowledge from the source domain. Theoretically, the characteristic kernel is the natural choice for MMD. However, in specific applications, non-characteristic counterparts might be more appropriate than characteristic ones [3]. Several MMD-based DA methods employ non-characteristic kernels (such as the linear kernel [16, 20, 25] and the polynomial kernel [1, 2]) or the non-kernel linear transformation [14,15,16, 20, 37] to cope with domain shift. Despite the state-of-the-art performance achieved by the non-characteristic kernel based methods, in this paper we point out that there is still much room to improve these current methods. To be specific, with a deep insight into the recent MMD methods, we can come to the conclusion that the non-characteristic kernel based MMD could lose some statistical information that is important for DA.
To capture more information about distributions, this paper designs a new distribution metric termed the maximum mean and covariance discrepancy (MMCD). This metric is designed to address both the first- and second-order statistical information in the RKHS. Specifically, MMCD is comprised of MMD and our proposed maximum covariance discrepancy (MCD). MCD evaluates the Hilbert–Schmidt norm of the difference between covariance operators such that it addresses the second-order statistics in the RKHS. Since MMCD unites MCD and MMD, MMCD is able to simultaneously consider the first- and second-order statistics in the RKHS. Thus, when the non-characteristic kernel is used, MMCD has the ability to capture more information about distributions than MMD does. This point will be analyzed in Sects. 3.2 and 3.3.
To verify the efficacy of MMCD, we propose an unsupervised DA method that is based on MMCD (McDA) in the joint distribution adaptation (JDA) paradigm [20]. JDA has been chosen because it jointly adapts both the marginal and conditional distributions across domains. It is not easy to optimize McDA due to its non-convex term. To address this issue, we approximate the non-convex term with its convex upper bound, thereby yielding a closed-form solution to each sub-problem. Experiments in cross-domain image classification on two datasets (PIE and Office-Caltech) show the effectiveness of McDA in the JDA paradigm when compared with the baseline methods, which implies the efficacy of MMCD.
The rest of this paper is organized as follows. Section 2 reviews the related works. Section 3 introduces the proposed maximum mean and covariance discrepancy. Section 4 details the MMCD based unsupervised DA model McDA, along with an optimization algorithm to solve it. In Sect. 5, we report the performance of McDA on two benchmark datasets. Finally, Sect. 6 concludes the paper.
2 Related Work
Domain adaptation (DA) has been widely studied in extensive literatures [26, 27]. DA can be categorized into two groups supervised DA and unsupervised DA according to whether or not the data in the target domain is labeled [4, 6, 25, 38, 40]. In this paper, we focus on the unsupervised setting. To tackle the unsupervised DA problem, many methods has been proposed including sample reweighting [17], projection learning [25] and subspace alignment [8, 36]. Among them, a popular way is to learn the domain-invariant representation which minimize the distribution discrepancy across domains. Notably, two methods of measuring the distribution discrepancy have been widely adopted in DA models: maximum mean discrepancy (MMD) metric [11] and the explicit order-wise distance between distributions.
As a representative metric used to measure distribution discrepancy, MMD has been widely used in DA methods. For instance, Pan et al. [25] developed transfer component analysis (TCA) to learn invariant features across domains in the reproducing kernel Hilbert space (RKHS). To investigate the benefits of conditional distributions, Long et al. [20] constructed a joint distribution adaptation (JDA) paradigm which adopted MMD in order to jointly evaluate the marginal and conditional distribution differences across domains. JDA yielded the state-of-the-art classification performance. Recently, Hsieh et al. [15] generalized JDA in order to tackle heterogeneous DA problems. Later, the closest common space learning [16] was proposed to deal with imbalanced cross-domain data problem in the frame of JDA. With advances in deep learning, MMD-based deep models have also made great success in adaptation performance across domains. Long et al. [19] proposed a deep adaptation network architecture, in which a multiple-kernel MMD was utilized in order to match cross-domain distributions of multiple task-specific CNN layers, so as to boost the transferability of deep features. Recently, Long et al. [21] have proposed a new DA method to be used in deep networks, i.e., residual transfer networks, which was able to jointly learn both adaptive classifiers and transferable features. To be different, Our MMCD explores both the first- and second-order statistics in the RKHS.
In addition to MMD-based models, recent work has explicitly matched the order-wise statistics of cross-domain distributions, thereby taking the statistics of distributions into account. For example, Sun et al. [35] proposed an efficient unsupervised DA method, which they designated as correlation alignment, to align the covariance matrix of the source and target distributions. Jiang et al. [18] proposed to use the second moment matching so as to learn the domain-invariant feature. Zong et al. [41] took a different approach which explores both the least-square regression and mean–covariance feature matching in order to enhance the performance of cross-corpus speech emotion recognition. Zellinger et al. [39] took a step forward to explore higher order moment which learns domain-invariant representations by means of explicitly order-wise moment matching. Different from the above mentioned methods which all study in the original feature space, our MMCD, however, explores the higher order statistics in the RKHS by using the kernel trick.
It is noted that MMCD is related to the work of [22] which embeds distributions in a finite dimensional feature space, and matches the mean and covariance feature statistics in order to train generative adversarial networks. The proposed MMCD differs from this approach in two ways. Firstly, MMCD function set is from the RKHS which is infinitely dimensional in the setting of PSD kernels. In comparison, the function set used by [22] is parameterized by a finite-dimensional CNN. Secondly, MCD can be represented as the Hilbert–Schmidt norm. By contrast, the covariance matching in [22] is related to the nuclear norm. Besides, we notice that the empirical estimator of squared MCD has similar form to that from [28, 29]. However, they are different in three aspects. Firstly, MCD is defined in the frame of integral probability metrics [24] and can be regarded as the second-order generation of MMD, while [28] uses the empirical estimator of squared MCD directly. Secondly, MCD serves as distribution metric, while [28] uses covariance operators as image representation. Thirdly, this paper unites both MMD and MCD as MMCD to measure the distribution discrepancy.
3 Maximum Mean and Covariance Discrepancy
This section will introduce the maximum covariance discrepancy (MCD) and its joint variant the maximum mean and covariance discrepancy (MMCD). In addition, several theoretical analyses will be provided. At first, we introduce the notations that are used throughout this section.
Let \(\mathcal{H}\) be a reproducing kernel Hilbert space (RKHS) over X with associated kernel \(k\left( { \cdot , \cdot } \right) \), whose canonical feature map is \(\phi \left( x \right) = k\left( {x, \cdot } \right) \) for \(x \in X\). Let x and y be random variables defined on X, we assume \(x\sim p\) and \(y\sim q\), where \(x\sim p\) indicates x follows distribution p. The notation \({E_{x\sim p}}\) denotes the expectation with respect to p, and we abbreviate \({E_{x\sim p}}\) and \({E_{x\sim q}}\) to \({E_x}\) and \({E_y}\) respectively when there is no ambiguity. Following [12], when given \(f,g \in \mathcal{H}\), the tensor product operator \(f \otimes g{:}\,\mathcal{H} \rightarrow \mathcal{H}\) is defined as \(\left( {f \otimes g} \right) h = f{\left\langle {g,h} \right\rangle _\mathcal{H}}\) for all \(h \in H\), where \( \otimes \) denotes the tensor product. Based on the tensor product operator and probability distributions, the centered covariance operator \(C{:}\,\mathcal{H} \rightarrow \mathcal{H}\) associated with the distribution p has the definition \(C\left[ p \right] = {E_{x\sim p}}\left[ {\phi \left( x \right) \otimes \phi \left( x \right) } \right] - {E_{x\sim p}}\left[ {\phi \left( x \right) } \right] \otimes {E_{x\sim p}}\left[ {\phi \left( x \right) } \right] \). At last, we make two assumptions in our paper. First, we assume the kernel \(k\left( { \cdot , \cdot } \right) \) to be bounded, which makes the covariance operator bounded and the mean embedding of distributions in \(\mathcal{H}\) exists [11]. Second, we assume \(\mathcal{H}\) to be separable such that the basis of \(\mathcal{H}\) is finite or countably infinite.
3.1 Definition
With the assumptions and notations above, we define the MCD in Definition 1.
Definition 1
The maximum covariance discrepancy (MCD) is defined as
where \(\left\{ {{e_i}|i \in I} \right\} \) is an orthogonal basis of \(\mathcal{H}\), \(\left\| a \right\| = {(\sum \nolimits _{i,j \in I} {a_{ij}^2} )^{1/2}}\), and the notation \({\mathop {\mathrm{cov}}} \) has the following formula: \({\mathop {\mathrm{cov}}} \left[ {{e_i}\left( x \right) ,{e_j}\left( x \right) } \right] = {E_x}\left[ {{e_i}\left( x \right) {e_j}\left( x \right) } \right] - {E_x}\left[ {{e_i}\left( x \right) } \right] {E_x}\left[ {{e_j}\left( x \right) } \right] \).
Next, we show that the (1) can be associated with the Hilbert–Schmidt norm with the following lemma:
Lemma 1
where \({\left\| {} \right\| _{{\mathrm{HS}}}}\) denotes the Hilbert–Schmidt norm of the vectors in \({\mathrm{HS}}\left( \mathcal{H} \right) \), which is the Hilbert space of Hilbert–Schmidt operators mapping from \(\mathcal{H}\) to \(\mathcal{H}\).
Proof
By using the reproducing property of the kernel k, we have
Similarly, we also have
In terms of (3) and (4), we have
where \({h^*}\) denotes the tensor product operator of h which belongs to the tensor product space \(\mathcal{H} \otimes \mathcal{H}\). Since \(\left\{ {{e_i}|i \in I} \right\} \) is the orthogonal basis of \(\mathcal{H}\), \(\left\{ {{e_i} \otimes {e_j}|i,j \in I} \right\} \) consists of the orthogonal basis of \(\mathcal{H} \otimes \mathcal{H}\). Thus, the second equality holds. The third equality follows from the following fact:
\(\square \)
To compute it, we express MCD as the expansion formula with respect to kernel functions, using the following lemma:
Lemma 2
The squared MCD terms of kernel functions is
where \(x'\) and \(y'\) are independent copy of x and y, with the same distribution, respectively.
Given the limited observations \(X = \left\{ {{x_1},\ldots ,{x_n}} \right\} \) and \(Y = \left\{ {{y_1},\ldots ,{y_m}} \right\} \) sampled from p and q, and based on Lemma 2 and statistical theory, an empirical estimator of squared MCD can be given by
where \({({K_{XX}})_{ij}} = k({x_i},{x_j})\), \({({K_{XY}})_{ij}} = k({x_i},{y_j})\), \({({K_{YY}})_{ij}} = k({y_i},{y_j})\), and \({L_n} = {I_n} - {\frac{1}{n}}1_n^T{1_n}\) wherein \({I_n}\) is the identity matrix of size n, and \({1_n}\) is the vector of ones with length n. This can be notated as, \({L_m} = {I_m} - {\frac{1}{m}}1_m^T{1_m}\). To be concise, we can rewrite (8) as the elegant formula
where
and
Moreover, we can also yield another equivalent representation of (8), as follows:
where \(k\left( {x,y} \right) = \phi {\left( x \right) ^T}\phi \left( y \right) \), wherein \(\phi \left( X \right) = \left[ {\phi \left( {{x_1}} \right) ,\ldots ,\phi \left( {{x_n}} \right) } \right] \) and \(\phi \left( Y \right) = \left[ {\phi \left( {{y_1}} \right) ,\ldots ,\phi \left( {{y_m}} \right) } \right] \). This form will be utilized in the later section.
Now, we unite MMD and MCD into a joint metric, namely maximum mean and covariance discrepancy (MMCD) in order to capture more statistical information from data distributions.
Definition 2
The maximum mean and covariance discrepancy (MMCD) is defined as
where \(\mu \left[ p \right] = {E_x}\left[ {\phi \left( x \right) } \right] \) and \(\beta \) is a non-negative parameter.
According to this definition, MMCD can be proven as a distribution metric when the associate kernel of \(\mathcal{H}\) is characteristic. This property can be established by Theorem 1.
Theorem 1
Let the associated kernel of \(\mathcal{H}\) be characteristic. Then \({\mathrm{MMCD}}\left[ {p,q,\mathcal{H}} \right] = 0\) if and only if \(p = q\). Moreover, \({\mathrm{MMCD}}\left[ {p,q,\mathcal{H}} \right] \) is a metric on the space of probability distribution.
Proof
According to [7], a metric d should satisfy four properties: non-negativity, \(d(p, q)=0 \Leftrightarrow p=q\), symmetry and triangle inequality. Apparently, MMCD is non-negative due to the non-negativity of the norm. For the necessity and sufficiency of the second property, first, under the condition \(p = q\), \({\mathop {\mathrm{MMCD}}\nolimits } \left[ {p,q,\mathcal{H}} \right] = 0\); conversely, \(p = q\) can be deduced according to the definition of characteristic kernel [9] and \({\mathop {\mathrm{MMD}}\nolimits } \left[ {p,q,\mathcal{H}} \right] = 0\). As a side assertion, it is easy to verify that \({\mathop {\mathrm{MMCD}}\nolimits } \left[ {p,q,\mathcal{H}} \right] = {\mathop {\mathrm{MMCD}}\nolimits } \left[ {q,p,\mathcal{H}} \right] \) which demonstrates the symmetric property of MMCD. The only property left to prove is the triangle inequality. First, we need to prove that the \({\mathrm{MCD}}\) meets this property. Let \({{\mathrm{d}}_{ij}}\left( {x,y} \right) = {\mathop {\mathrm{cov}}} [ {{e_i}\left( x \right) ,{e_j}\left( x \right) } ] - {\mathop {\mathrm{cov}}} [ {{e_i}\left( y \right) ,{e_j}\left( y \right) } ]\), and then we have
where \(z\sim r\). Next, we prove the triangle inequality holds for the \({\mathrm{MMCD}}\). To simplify the notation, let \({\mathop {\mathrm{M}}\nolimits } \left[ {p,q} \right] = {\mathop {\mathrm{MMD}}\nolimits } \left[ {p,q,\mathcal{H}} \right] \), \(\mathrm{C}\left[ {p,q} \right] = {\mathop {\mathrm{MCD}}\nolimits } \left[ {p,q,\mathcal{H}} \right] \) and \(\mathrm{MC}\left[ {p,q} \right] = {\mathop {\mathrm{MMCD}}\nolimits } \left[ {p,q,\mathcal{H}} \right] \), and then we have
where the first inequality holds because both \({\mathrm{MMD}}\) and \({\mathrm{MCD}}\) meet the triangle inequality, and the second inequality holds from the Cauchy–Schwarz inequality. Taking the square root of both sides, there holds \({\mathop {\mathrm{MMCD}}\nolimits } \left[ {p,q} \right] \le {\mathop {\mathrm{MMCD}}\nolimits } \left[ {p,r} \right] + {\mathop {\mathrm{MMCD}}\nolimits } \left[ {r,q} \right] \). Obviously, \({\mathrm{MMCD}}\) meets the metric definition, and is, therefore, a metric. \(\square \)
In fact, while Theorem 1 relies on the condition that the kernel is characteristic, even if the condition does not hold, MMCD is still a pseudo-metric. According to (9) and [11], the empirical estimator of the squared MMCD can be given by
and
where
3.2 Explicit Representation of MMCD
As stated above, we have defined MMCD as the (pseudo-) metric of distributions. However, it is not easy to understand what specific information about the distribution is captured by MMCD. Towards this goal, we deduce the explicit representation of MMCD with the polynomial kernel and the linear kernel to illustrate the mechanism of MMCD.
We first introduce the explicit representation of MMCD associated with the polynomial kernel with a specific degree d, i.e., \(k\left( {x,y} \right) = {( {x^T{y} + c} )^d}\). According to the explicit feature map of the polynomial kernel [23, 31], MMCD can be explicitly written as
where
and \(C_d^i\) denotes the binomial coefficient, vec() converts the matrix into a column vector, and \( \otimes ^ix \) denotes the ith order tensor product of x, wherein \(x \sim p\).
By setting \(d=1\), (19) becomes
where \(x \sim p\) and \(y \sim q\). Given limited X and Y sampled from p and q, respectively, there is
where \({\mu _p} = \frac{1}{n}X{1_n}\) and \({\varSigma _p} = \frac{1}{n}X{H_n}{X^T}\) are mean vector and covariance matrix of X, respectively. Specially, when \(c=0\), the polynomial kernel becomes the linear kernel \(k\left( {x,y} \right) = x^T{y}\). Accordingly, (21) is exactly the explicit representation of MMCD with the linear kernel, and (22) is still the corresponding empirical estimator. As a result, both (21) and (22) show that MMD and MCD of MMCD measures the difference between means and covariances of distributions, respectively, when the polynomial kernel with degree \(d=1\) or the linear kernel is adopted.
Moreover, by setting \(d = 2\), there is
Combination of (19) with (23) shows that the MMD term of MMCD measures the difference between both the first and second raw moments of two distributions, whereas the MCD term measures the difference between covariances of up to the second raw moment of two distributions. An analogy for setting \(d>2\) means that the MCD term can capture higher order statistical information. From the statistical perspective, more insights into MCD remain to be further investigated in the future work.
3.3 Toy Examples
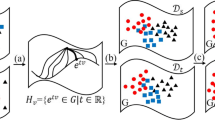
To clarify the efficacy of MMCD, which fuses both first- and second-order statistics in the RKHS, we illustrate two toy examples in Fig. 1a, b. We synthesize two groups of data points which follow different distributions in the two-dimensional space. Then, we fix one group of data points (in blue) and adjust the points from the other group (in red) by minimizing the values of MMD, MCD and MMCD via the gradient descent algorithm. Two types of kernels, that is, the linear kernel (Fig. 1a) and the polynomial kernel with \(d=2\) and \(c=1\) (Fig. 1b), are adopted. For concise, the details of computing their gradients are left in “Appendix A”.
Two toy examples for comparison of MMD, MCD and MMCD on the synthetic data. a Blue and red points are sampled from two Gaussian distributions whose both the mean and covariance are different, and the linear kernel is used, b blue points are sampled from a complicated distribution with the zero mean (0, 0), while red points are sampled from a Gaussian distribution with the mean (0, 1). The polynomial kernel with degree two is used therein. (Color figure online)
From Fig. 1a, through the distribution matching of MMD, the means of the red and blue points are almost identical, but the corresponding spread of these data points has different shapes. That is, MMD cannot match the covariances of two distributions. But, for MCD, the result is reversed, namely, the covariances of two groups of data points appear to be similar, but the corresponding means of these data points keep invariant as before and still differ from each other. Notably, after MMCD performs the distribution matching over two groups of data points, the distribution regions of both the red and blue points overlap highly. This is because MMCD simultaneously takes both mean and covariance differences into consideration, while either MMD or MCD could singly consider one aspect where the linear kernel is used.
Figure 1b displays, for the complicated distribution, the distribution matching results of MMD, MCD and MMCD, when the polynomial kernel with degree two is equipped. In this case, MMD induces different spread shapes, which corresponds to the high-order statistics. Meanwhile, it can be verified that the mean of the red data points is above that of the blue points which is zero. Thus, both MMD and MCD fail to make two distributions matched in this example. By using MMCD, the red and blue points become highly overlapped. This implies the efficacy of MMCD which explores both first- and second-order statistics in the RKHS, as compared to MMD and MCD. Hence, in contrast with MMD and MCD, MMCD has the promising potential of matching data distributions across domains for domain adaptation. More empirical analyses about the efficacy of MMCD in domain adaptation are shown in experiments.
4 Domain Adaptation Via MMCD
In this section, we apply the MMCD to unsupervised domain adaptation problem to verify the efficacy of MMCD. Unsupervised domain adaptation is still very challenging, as there is no supervised knowledge of target domains. Recently, joint distribution adaptation (JDA) [20] has been proven a promising domain adaptation method which matches both marginal and conditional distributions. JDA adopts MMD as the distance measurement of distributions; however, in the case of non-characteristic kernels, MMD based JDA loses the high-order statistical information. As mentioned in the previous section, MMCD has a better chance to capture more information about distributions than MMD. We thus substitute our MMCD for MMD in the frame of JDA to produce a new unsupervised domain adaptation method called McDA. In the subsection, we first introduce the problem formulation and notation, and then propose the McDA.
4.1 Problem Formulation and Notations
Given the dataset \({D_s} = \left\{ {{x_1},\ldots ,{x_{{n_s}}}} \right\} \) in the source domain with labels \({y_1},\ldots ,{y_{{n_s}}}\) and the unlabeled dataset \({D_t} = \left\{ {{x_{{n_s} + 1}},\ldots ,{x_{{n_s} + {n_t}}}} \right\} \) in the target domain under the assumption that both the marginal and conditional probability distributions in two domains are different, i.e., \({P_s}\left( {{x_s}} \right) \ne {P_t}\left( {{x_t}} \right) \) and \({P_s}\left( {{y_s}|{x_s}} \right) \ne {P_t}\left( {{y_t}|{x_t}} \right) \). We will adapt both marginal and conditional distributions in order to train a robust classifier on target dataset by leveraging the labeled source dataset.
For clarity, we now summarize several commonly used notations. The source and target data are denoted as \({X_s} = \left[ {{x_1},\ldots ,{x_{{n_s}}}} \right] \) and \({X_t} = \left[ {{x_{{n_s} + 1}},\ldots ,{x_{{n_s} + {n_t}}}} \right] \), respectively, and, for brevity, \(X = \left[ {{X_s},{X_t}} \right] \). The centeralized matrix \({L_n}\) is defined as \({L_n} = {I_n} - \frac{1}{n}1_n^T{1_n}\), where \({I_n}\) is the identity matrix of size n, and \({1_n}\) is the vector of ones with length n.
Since our model McDA is based on the JDA paradigm, it is necessary to review it before our text. JDA is to adapt both marginal and conditional distributions across domains by minimizing the following objective:
According to [20], \(\phi \) can serve as a linear transformation, or a non-linear feature map associated with the kernel. The property of MMD still is an open problem when substituting its feature map for linear transformation. Then we roughly regard the terms of (24) as the estimation of the generalized MMD whose feature map can be replaced with any arbitrary maps. Thus, following [20], we obtain two variants of our McDA according to the used feature maps: the linear transformation and the kernel feature map.
4.2 McDA
For the first variant of McDA, we adopt the linear transformation denoted by a matrix \(A \in {R^{m \times k}}\) to MMCD. According to (17), the distance between marginal distributions across domains can be converted into
where \({M_0}\) is defined as:
and \({Z_0}\), termed as the MCD matrix, is defined as:
Similarly, the discrepancy between conditional distributions across transformed domains can be cast as:
where \(c \in \left\{ {1,\ldots ,C} \right\} \), and \({M_c}\) is
wherein \({D_{s,c}}\left( {{D_{t,c}}} \right) = \left\{ {{x_i}|{x_i} \in {D_s}\left( {{D_t}} \right) ,{y_i} = c} \right\} \) and \({n_{s,c}}\left( {{n_{t,c}}} \right) = \left| {{D_{s,c}}\left( {{D_{t,c}}} \right) } \right| \), while \({Z_c}\) is defined as
By combining (25) and (28) and denoting \(H = {L_{{n_s} + {n_t}}}\), we can obtain the overall objective function as follows:
where I is the identity matrix of size k, a constraint is imposed to avoid yielding a trivial solution and the parameter \(\lambda \) remains (31) as a well-defined optimization problem.
For the second variant of McDA, we, however, adopt the kernel feature map \(\varphi {:}\,x \rightarrow \varphi \left( x \right) \) to MMCD. Let the kernel matrix \(K = \varphi {(X)^T}\varphi (X)\). The MMCD-based distance of both marginal and conditional cross-domain distributions is defined as:
Following [25], we employ the empirical kernel mapping \(K = (K{K^{ - 1/2}})({K^{ - 1/2}}K)\) and further introduce the low dimensional transformation in order to obtain \({\tilde{K}} = (K{K^{ - 1/2}}{\tilde{A}})({{\tilde{A}}^T}{K^{ - 1/2}}K) = KA{A^T}K\), where \(A = {K^{ - 1/2}}{\tilde{A}}\). Thus, substituting \({\tilde{K}}\) into (32) leads to the following minimization problem
4.3 Optimization Algorithm
Both (31) and (33) possess similar optimization problems; hence, only the optimization algorithm for (31) is provided here. Obviously, (31) is non-convex with the variable A and hard to optimize, as it contains a non-convex fourth-order term which is the second term of (31). In [20, 25], the optimization problem have the closed-form solution by solving a generalized eigen-decomposition problem. To preserve this property, we can approximate the fourth-order term in (31) with its convex upper bound by using the following theorem:
Theorem 2
The following inequality holds
where k is the reduced dimensionality and \(\sigma = {\left\| {{{(XH{X^T})}^{ - 1/2}}} \right\| ^2}\).
Proof
The first equation holds because \(XH{X^T}\) is semi-definite positive, while the second and forth inequalities follow the Cauchy–Schwarz inequality. In terms of the constraint of (31), \(k = Tr\left( {{A^T}XH{X^T}A} \right) = Tr\left( {{I_k}} \right) \). \(\square \)
According to Theorem 2, we absorb the constant of (34) in order to obtain the convex objective of (31), as follows:
Then, we derive the Lagrange function of (36), as follows:
where the diagonal matrix \(\varPhi \) denotes the Lagrange multipliers. By setting the derivative of (37) over A to zero, we can obtain the following generalized eigenvalue decomposition problem:

Following the JDA paradigm [20], we learn the transformation matrix via (38) and can then project all the samples to a low-dimensional subspace. Based on the labeled source data, we can train a specific classifier to assign pseudo-labels to the samples of the target domain, and then repeat the procedure above until convergence. The overall procedure of McDA is summarized in Algorithm 1.
5 Experiments
This section is to verify the effectiveness of McDA by comparing its classification performance against the baseline methods on two benchmark datasets including PIE and Office-Caltech.
5.1 Datasets
The PIE dataset contains face images of size 32x32 from 68 individuals with different poses, illuminations and expressions. Following [20], We evaluate our method using five subsets from the PIE face dataset. Each subset corresponds to a different pose, e.g. P1 (left), P2 (upward), P3 (downward), P4 (frontal) and P5 (right). We then construct 20 cross-domain datasets via a pairwise combination of subsets, i.e., P1 \(\rightarrow \) P2, P1 \(\rightarrow \) P3, \( \ldots \), P5 \(\rightarrow \) P4. Since the source and target face images from each cross-domain dataset have different poses, they will follow different distributions.
Office-Caltech is a widely used dataset for domain adaptation which contains four domains: A (Amazon), W (Webcam), D (DSLR) and C (Caltech-256) [13, 30]. The SURF feature is extracted for each image before being converted into histograms over an 800-bin codebook clustered by k-means on the Amazon database. We construct 12 cross-domain datasets by pairwise combination of the four domains, i.e., \(\hbox {C}\rightarrow \hbox {A}\), \(\hbox {C}\rightarrow \hbox {W}\), \( \ldots \), \(\hbox {D}\rightarrow \hbox {W}\). Several sample images from PIE and Office-Caltech datasets are illustrated in Fig. 2.
5.2 Cross-Domain Image Classification
We compare McDA with five typical and related baseline methods including nearest neighbor classifier (NN), principal component analysis (PCA), correlation alignment (CA) [35], transfer component analysis (TCA) [25], geodesic flow kernel (GFK) [10], and joint domain adaptation (JDA) [20]. As suggested by [10, 20], NN serves as the base classifier for both McDA and compared methods.
We follow the evaluation protocol of [20] for both benchmark datasets. In order to give a fair comparison, we also give the candidate ranges for the parameters used in the compared methods. Both TCA and JDA have a regularization parameter, and its candidate values are as follows: {0.001, 0.01, 0.1, 1, 10, 100}. For McDA, it is rather expensive to search all the possible parameter combinations for the best average accuracy. Since McDA shares the reduced dimensionality k and the regularization parameter \(\lambda \) with JDA, we directly set \(\lambda \) in McDA to the specific value at which JDA achieves its best accuracy and then search the other parameter \(\beta \) from {0.001, 0.01, 0.1, 1, 10, 100} at each k. We further empirically set the number of iterations N in McDA to 10 to guarantee convergence. For the compared subspace learning methods, the subspace dimensionality ranges from {10, 20, \(\ldots \), 200}. For TCA, JDA and McDA, we apply the linear transformation for the PIE dataset and the linear kernel for the Office-Caltech dataset as suggested by [20]. We adopt the broadly used classification accuracy on target dataset as the evaluation metric.
Figure 3 shows the compared classification accuracy versus different subspace dimensionalities. For two benchmark datasets, McDA is superior to all the compared methods in all dimensionalities in terms of average accuracy. The average accuracy is defined as the mean of classification accuracy over different cross-domain datasets.
Table 1 illustrates the accuracy of facial image classification on 20 cross-domain datasets of the PIE dataset. The results of the experiment show that our method outperforms the baseline methods in quantity. Importantly, McDA exceeds the average accuracy of JDA by 9.7%. Table 2 reports object recognition accuracy when the Office-Caltech dataset is used. McDA also outweighs the baseline methods in most situations and its average accuracy is higher than those of the compared baseline methods. Based on the analysis above, the prominent performance of McDA implies the efficacy of the proposed MMCD.
5.3 Sensitivity Analysis
5.3.1 Kernel Choice
It is an open problem to choose suitable kernels for kernel-based learning methods. To investigate the effect of the kernels on the performance of McDA, we run McDA and the baseline JDA on two datasets by comparing various kernel and non-kernel based cases. For the kernel based cases, the linear kernel, the polynomial kernel of degree two \({( {x^T{y} + 1} )^2}\), the Gaussian kernel \(\exp ( { - \left\| {x - y} \right\| _2^2/{2\sigma ^2} } )\), and the exponential kernel \(\exp ( { - \left\| {x - y} \right\| _2/\sigma } )\) are used. For the non-kernel case, the linear transformation in the original space is performed. For both Gaussian and exponential kernels, \(\sigma \) is set to the median of pairwise distances between all the samples. For the PIE dataset, it is hard to perform the eigenvalue decomposition over the large kernel matrix for the sake of large sample sizes. A compromise solution is to construct a small dataset termed PIE-sub, which is a subset of the PIE dataset by randomly selecting 10 images from each class. As a result, each domain of PIE-sub consists of 680 images from 68 classes.
Table 3 shows that McDA consistently outperforms the baseline method JDA in all the cases. This implies that the MCD regularization term in McDA is beneficial for capturing more distribution information so as to promote the performance of domain adaptation in a variety of kernels. It is worth mentioning that in non-kernel condition the performance of McDA is also enhanced due to the presence of the MCD term.
5.3.2 Parameter Selection
Our McDA has two regularization parameters: \(\lambda \) which is to avoid the optimization problem to be ill-defined, and \(\beta \) which is to balance the importance of the MCD term. In order to analyze the effect of parameters on the performance, we vary \(\lambda \) and \(\beta \) from the range {\(10^{-3}\), \(10^{-2.5}\), ..., \(10^2\)} and run McDA on PIE and Office-Caltech datasets. The reduced dimensionality is set to the optimal value according to Fig. 3, i.e., \(k=90\) for PIE and \(k=20\) for Office-Caltech. Figure 4a shows that small values of \(\lambda \) and relatively big values of \(\beta \) for the PIE dataset help achieve high accuracy. This could result from both significant mean and covariance differences between source and target data distributions. It can be seen from Fig. 4b that the performance of McDA is relatively stable to the change of the values of \(\lambda \) and \(\beta \) in the area of \(\lambda \ge \beta \) on the Office-Caltech dataset. Hence, \(\lambda \in \left[ {0.001,1} \right] \) and \(\beta \in \left[ {0.01,10} \right] \) are suggested for accomplishing enhanced performance.
6 Conclusion
This paper proposes a new distribution metric namely maximum mean and covariance discrepancy (MMCD) which unites MMD with the proposed maximum covariance discrepancy (MCD). MMCD is able to capture more information about distributions compared to MMD. Based on MMCD, we developed a new domain adaptation method in the joint distribution adaptation paradigm. Experiments conducted on two benchmark datasets verify the effectiveness of our method, which implies the efficacy of MMCD. In the future, we plan to apply the MMCD to other domain adaptation methods and test them on various applications.
References
Baktashmotlagh M, Harandi M, Salzmann M (2016) Distribution-matching embedding for visual domain adaptation. J Mach Learn Res 17(1):3760–3789
Baktashmotlagh M, Harandi MT, Lovell BC, Salzmann M (2013) Unsupervised domain adaptation by domain invariant projection. In: IEEE international conference on computer vision, pp 769–776
Borgwardt KM, Gretton A, Rasch MJ, Kriegel HP, Schölkopf B, Smola AJ (2006) Integrating structured biological data by kernel maximum mean discrepancy. Bioinformatics 22(14):e49–e57
Bruzzone L, Marconcini M (2010) Domain adaptation problems: a DASVM classification technique and a circular validation strategy. IEEE Trans Pattern Anal Mach Intell 32(5):770–787
Cao X, Wipf D, Wen F, Duan G, Sun J (2013) A practical transfer learning algorithm for face verification. In: IEEE international conference on computer vision, pp 3208–3215
Dai W, Yang Q, Xue GR, Yu Y (2007) Boosting for transfer learning. In: Ghahramani Z (ed) Proceedings of the 24th international conference on machine learning. ACM, New York, pp 193–200
Dudley R, Fulton W, Katok A, Sarnak P, Bollobás B, Kirwan F (2002) Real analysis and probability. Cambridge University Press, Cambridge. https://doi.org/10.1017/CBO9780511755347
Fernando B, Habrard A, Sebban M, Tuytelaars T (2013) Unsupervised visual domain adaptation using subspace alignment. In: IEEE International conference on computer vision, pp 2960–2967
Fukumizu K, Gretton A, Sun X, Schölkopf B (2008) Kernel measures of conditional dependence. In: Advances in neural information processing systems, pp 489–496
Gong B, Shi Y, Sha F, Grauman K (2012) Geodesic flow kernel for unsupervised domain adaptation. In: IEEE international conference on computer vision, pp 2066–2073
Gretton A, Borgwardt KM, Rasch MJ, Schölkopf B, Smola A (2012) A kernel two-sample test. J Mach Learn Res 13(Mar):723–773
Gretton A, Bousquet O, Smola A, Schölkopf B (2005) Measuring statistical dependence with Hilbert–Schmidt norms. In: International conference on algorithmic learning theory, pp 63–77
Griffin G, Holub A, Perona P (2007) Caltech-256 object category dataset. Technical Report 7694, California Institute of Technology
Guo Y, Ding G, Liu Q (2015) Distribution regularized nonnegative matrix factorization for transfer visual feature learning. In: ACM international conference on multimedia retrieval, pp 299–306
Hsieh YT, Tao SY, Tsai YHH, Yeh YR, Wang YCF (2016) Recognizing heterogeneous cross-domain data via generalized joint distribution adaptation. In: IEEE international conference on multimedia and expo, pp 1–6
Hsu TH, Chen W, Hou C, Tsai YH, Yeh Y, Wang YF (2015) Unsupervised domain adaptation with imbalanced cross-domain data. In: IEEE international conference on computer vision, pp 4121–4129
Huang J, Smola AJ, Gretton A, Borgwardt KM, Schölkopf B (2006) Correcting sample selection bias by unlabeled data. In: Advances in neural information processing systems, pp 601–608
Jiang W, Deng C, Liu W, Nie F, Chung F, Huang H (2017) Theoretic analysis and extremely easy algorithms for domain adaptive feature learning. In: Proceedings of the international joint conference on artificial intelligence, pp 1958–1964
Long M, Cao Y, Wang J, Jordan M (2015) Learning transferable features with deep adaptation networks. In: International conference on machine learning, pp 97–105
Long M, Wang J, Ding G, Sun J, Yu PS (2013) Transfer feature learning with joint distribution adaptation. In: IEEE international conference on computer vision, pp 2200–2207
Long M, Zhu H, Wang J, Jordan MI (2016) Unsupervised domain adaptation with residual transfer networks. In: Advances in neural information processing systems, pp 136–144
Mroueh Y, Sercu T, Goel V (2017) McGan: mean and covariance feature matching GAN. In: International conference on machine learning, pp 2527–2535
Muandet K, Fukumizu K, Sriperumbudur B, Schölkopf B et al (2017) Kernel mean embedding of distributions: a review and beyond. Found Trends Mach Learn 10(1–2):1–141
Müller A (1997) Integral probability metrics and their generating classes of functions. Adv Appl Probab 29(2):429–443
Pan SJ, Tsang IW, Kwok JT, Yang Q (2011) Domain adaptation via transfer component analysis. IEEE Trans Neural Netw 22(2):199–210
Pan SJ, Yang Q (2010) A survey on transfer learning. IEEE Trans Knowl Data Eng 22(10):1345–1359
Patel VM, Gopalan R, Li R, Chellappa R (2015) Visual domain adaptation: a survey of recent advances. IEEE Signal Process Mag 32(3):53–69
Quang M.H, San Biagio M, Murino V (2014) Log-Hilbert–Schmidt metric between positive definite operators on Hilbert spaces. In: Advances in neural information processing systems, pp 388–396
Quang Minh H, San Biagio M, Bazzani L, Murino V (2016) Approximate log-Hilbert–Schmidt distances between covariance operators for image classification. In: IEEE conference on computer vision and pattern recognition, pp 5195–5203
Saenko K, Kulis B, Fritz M, Darrell T (2010) Adapting visual category models to new domains. In: European conference on computer vision, pp 213–226
Schölkopf B, Smola AJ, Bach F et al (2002) Learning with kernels: support vector machines, regularization, optimization, and beyond. MIT Press, Cambridge
Shen J, Qu Y, Zhang W, Yu Y (2018) Wasserstein distance guided representation learning for domain adaptation. In: AAAI conference on artificial intelligence
Si S, Tao D, Geng B (2010) Bregman divergence-based regularization for transfer subspace learning. IEEE Trans Knowl Data Eng 22(7):929–942
Sriperumbudur BK, Gretton A, Fukumizu K, Lanckriet GRG, Schölkopf B (2008) Injective Hilbert space embeddings of probability measures. In: Annual conference on learning theory, pp 111–122
Sun B, Feng J, Saenko K (2016) Return of frustratingly easy domain adaptation. In: AAAI conference on artificial intelligence, pp 2058–2065
Sun H, Liu S, Zhou S (2016) Discriminative subspace alignment for unsupervised visual domain adaptation. Neural Process Lett 44(3):779–793
Wang T, Ye T, Gurrin C (2016) Transfer nonnegative matrix factorization for image representation. In: International conference on multimedia modeling, pp 3–14
Xie X, Sun S, Chen H, Qian J (2018) Domain adaptation with twin support vector machines. Neural Process Lett 48(2):1213–1226
Zellinger W, Grubinger T, Lughofer E, Natschläger T, Saminger-Platz S (2017) Central moment discrepancy (CMD) for domain-invariant representation learning. In: International conference on learning representations
Zhu L, Zhang X, Zhang W, Huang X, Guan N, Luo Z (2017) Unsupervised domain adaptation with joint supervised sparse coding and discriminative regularization term. In: IEEE international conference on image processing, pp 3066–3070
Zong Y, Zheng W, Zhang T, Huang X (2016) Cross-corpus speech emotion recognition based on domain-adaptive least-squares regression. IEEE Signal Process Lett 23(5):585–589
Acknowledgements
This work was supported by the National Natural Science Foundation of China (61806213, 61702134, U1435222).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A: Gradient Computation
Appendix A: Gradient Computation
According to (16), when the polynomial kernel of degree d is adopted, the gradient of the empirical estimator of squared MMD with respect to the data matrix \(A=[X, Y]\) is given by
where \({\left( {{K_{d - 1}}} \right) _{ij}} = {\left( {{A_i^T}A_j + c} \right) ^{d - 1}}\) and \( \circ \) denotes the element-wise product. Likewise, there holds
and
The gradients of MMD, MCD and MMCD with the linear kernel can be obtained by setting \(d=1\) and \(c=0\) in (39)–(41), respectively.
Rights and permissions
About this article
Cite this article
Zhang, W., Zhang, X., Lan, L. et al. Maximum Mean and Covariance Discrepancy for Unsupervised Domain Adaptation. Neural Process Lett 51, 347–366 (2020). https://doi.org/10.1007/s11063-019-10090-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11063-019-10090-0