Abstract
We previously demonstrated that the acquired resistance because of Hsp27 activation weakens the cytotoxic effect of t-AUCB on glioblastoma cells. Since autophagy is regarded as a survival mechanism for cells exposed to cytotoxic agents, the aim of this study is to investigate whether t-AUCB induces autophagy and whether Hsp27 and autophagy are interacted with each other. Our data demonstrated that t-AUCB induces autophagy in glioblastoma cells and regulates multiple autophagy related-gene expression. t-AUCB induces overexpression of Atg7, which is downstream of Hsp27 and participates in the resistance of glioblastoma cells to t-AUCB treatment. Hsp27 inhibitor quercetin suppresses Atg7 expression and strengthens t-AUCB-induced cell death by autophagy blockage. We concluded that combination of quercetin and t-AUCB might be a potential strategy for glioblastoma treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Our previous studies have demonstrated that, soluble epoxide hydrolase inhibitor (sEHi) t-AUCB inhibits human glioblastoma cell growth, induces G1 cell cycle arrest and p38 MAPK/Hsp27 activation which confer apoptosis resistance [1–3]. The cytotoxic effect of t-AUCB on glioblastoma cells is certain, but multiple mechanisms may participate in treatment resistance, such as p38 MAPK/Hsp27 activation that is known by us. Thus, to further investigate unknown mechanisms that may initiate t-AUCB treatment resistance is necessary.
Autophagic process is characterized by cellular self-digestion and the removal of excessive or dysfunctional organelles and proteins [4–6]. Autophagy is either a physiological process in normal cells or a survival mechanism under special conditions, such as hypoxia, stress or nutrient deprivation [7]. Since autophagy participates in multiple key biological processes, drugs that can modulate autophagy have been developed for therapeutic purposes [7–9]. Recently, more and more studies demonstrated that, autophagy induces chemo- or radio-resistance and may be a potential anticancer therapeutic target [10–12], although the mechanism of autophagy influenced resistance is still unclear. In the present study, we investigate whether t-AUCB treatment induces autophagy, and whether Hsp27 and autophagy are interacted with each other.
Materials and Methods
Reagents
The sEH inhibitor t-AUCB was granted from Professor Bruce D. Hammock (Department of Entomology and UCD Cancer Research Center, University of California, Davis, CA, USA) [13]. The Hsp27 inhibitor quercetin was purchased from Sigma-Aldrich (St. Louis, MO, USA). All of agents were dissolved in dimethyl sulfoxide (DMSO). The concentration which was never exceeded 0.1 % (v/v) was diluted in serum-supplemented medium immediately before use.
Cell culture
Human glioblastoma cell lines U251 and U87 were provided by ATCC (American Type Culture Collection) as we described previously [2]. All cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10 % fetal bovine serum (FBS) and 1 % penicillin and streptomycin (complete medium). Cells were maintained at 37 °C in a humidified atmosphere of 95 % air and 5 % CO2.
Cell growth assay
Cell growth ability was tested by using cell counting kit-8 (CCK-8) from Dojindo Laboratories (Kumamoto, Japan) following the manufacturer’s instructions. Briefly, cells were transplanted into a 96-well plate with density of 5000 cells/well and then treated differently. Cells were cultured in humidified incubator containing 5 %CO2 and 95 % air. 48 h later, culture medium in each well was discarded, and 100 μl fresh serum-free medium contained 10 μl CCK-8 solution was added into each well. After 2-h incubation, the optical density value (absorbance) was recorded at 450 nm using an enzyme-linked immunosorbent assay plate reader (Bio-Rad Laboratories, Inc., Berkeley, CA, USA).
Electron microscopy
Cells were differently treated and then the electron microscopy was performed. Briefly, after treatment, cells were harvested, pelleted and fixed with 2.5 % glutaraldehyde and then postfixed with 1 % osmium tetraoxide, dehydrated in a graded series of ethanol concentrations and then embedded in Epon-Araldite resin. The ultrathin sections were mounted, stained with lead citrate and then examined on a JEOL JEM-1011 transmission electron microscope. The number of autophagic vacuoles was determined manually for a minimum of 50 cells for each sample and the photos were taken.
RNA extraction and quantitative real-time PCR array analysis
Total RNA was extracted from cells lysated by TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. The quality and quantity of the RNA purity were assessed by spectrophotometer and standard electrophoresis. The RNA samples were tested for the relative expression of genes involved in autophagy by using RT2 Profiler™ PCR Array Human Autophagy (Qiagen) according to the manufacturer’s instructions. Briefly, the RNA was reverse transcribed to form cDNA by using SuperScript III Reverse Transcriptase (Invitrogen). Samples were diluted in SuperArray PCR master mix (RT2 SYBR Green, Qiagen) according to the supplier’s directions and pipetted into 96-well PCR array plates. Real-time PCR was performed in technical duplicates (ABI Prism 7900HT Sequence Detection System; Applied Biosystems). For data analysis, the ΔΔCt (comparative threshold cycle) method was used. Firstly, calculate the ΔCt for each pathway-focused gene in each treatment group. ΔCt (group1) = average Ct—average of HK genes’ Ct for group1 array; ΔCt (group2) = average Ct—average of HK genes’ Ct for group2 array. Secondly, calculate the ΔΔCt for each gene across two groups, ΔΔCt = ΔCt (group2)—ΔCt (group1), where group1 is the control and group2 is the experimental. Then, calculate the fold-change for each gene from group1 to group2 as \(2^{{ - \Delta \Delta {\text{C}}_{\text{t}} }}\). The average expression of housekeeping genes (β-actin) was used for normalization of the data.
RNA interference
Cells were grown in 10-cm culture dish and transiently transfected with Hsp27 or Atg7 specific small interfering RNA (siRNA). The siRNA oligos of Hsp27 (sc-29350), Atg7 (sc-41447) and the negative control siRNAs were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Lipofectamine RNAiMAX Transfection Reagent (Invitrogen) was used for siRNA transfection according to the manufacturer’s protocol.
Western blot analysis
Cell protein was lysates in ice-cold RIPA buffer (Beyotime Institute of Biotechnology, Shanghai, China) containing with Phenylmethanesulfonyl fluoride (PMSF) and protease inhibitor cocktail. The whole cell lysates were separated by SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to a polyvinylidene fluoride membrane (Millipore Corporation, Bedford, MA, USA). All membranes were probed with primary antibodies after 4 °C overnight, and followed by incubation with secondary antibody. Proteins were visualized with chemiluminescence luminol reagents (Beyotime Institute of Biotechnology, Shanghai, China). Antibodies against β-actin (#3700), Atg7 (#8558) and Hsp27 (#2402) were purchased from Cell Signaling Technology (Beverly, MA, USA). Antibody against LC3 (#AL221) was purchased from Beyotime Institute of Biotechnology (Shanghai, China). Public software ImageJ (National Institutes of Health, USA) was used to quantify the densitometry of the immunoblotting bands.
Statistical analysis
All experiments were replicated in triplicate at least. The SPSS version 17.0 software (SPSS Inc., Chicago, IL, USA) was applied for statistical analysis. Comparisons between treated groups and vehicle control were performed using independent t test, and expressed as mean ± standard deviation (SD). P < 0.05 were considered statistically significant.
Results
t-AUCB induces autophagy in glioblastoma cells
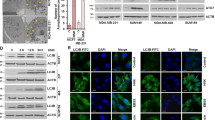
To investigate whether t-AUCB induce autophagy, U251 and U87 cells were treated with DMSO (vehicle control) or 200 μM t-AUCB for 24 h, and then analyzed by transmission electron microscopy. Membrane-bound vacuoles, the characteristic of autophagosomes, were observed in the cytoplasm of t-AUCB treated cells (Fig. 1a). As the computation shown, t-AUCB treatment initiates autophagy in U251 and U87 cells (Fig. 1b). The LC3 transforming from LC3-I to LC3-II is known to be important for autophagosome formation. We detected the LC3-I and LC3-II expression in U251 and U87 cells by western blotting. 200 μM t-AUCB enhanced the turnover from LC3-I to LC3-II (Fig. 1c).
t-AUCB induces autophagy in glioblastoma cells. a U251 and U87 cells were treated with DMSO (vehicle control) or 200 μM t-AUCB for 24 h, and then analyzed by transmission electron microscopy. Black arrows indicate the autophagic vacuoles. b The computation of the autophagic vacuoles per cell (*P < 0.05). c The LC3-I and LC3-II expression in U251 and U87 cells by western blotting. 200 μM t-AUCB enhanced the turnover from LC3-I to LC3-II. β-actin served as loading control
To further study the mechanisms, we analyzed the genes related to autophagy by RT2 Profiler™ PCR Array. U87 cells were treated with DMSO (vehicle control) or 200 μM t-AUCB for 24 h, and then the PCR array was performed. The Supplementary Table listed all analyzed genes and the alteration of the autophagy related-gene expression profile. Gene expression level changes that were more than 2-fold in t-AUCB-treated versus untreated U87 cells were listed in Table 1.
Atg7 is downstream of Hsp27 and participates the resistance of glioblastoma cells to t-AUCB
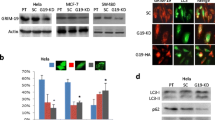
As the PCR results shown, Atg7 expression was upregulated in t-AUCB treated cells. We then tested the protein expression of Atg7 by western blotting. The results showed that 200 μM t-AUCB significantly increases Atg7 expression (Fig. 2a). Atg7 plays a key role in autophagosome formation [14]. Autophagy is considered to be associated with cancer resistance [10]. We previously reported that Hsp27 confers resistance of glioblastoma cells to t-AUCB [2]. Atg7 has been reported to be downstream of Hsp27 by others [15]. Thus, we hypothesized that Atg7 may be regulated by Hsp27 and involved in t-AUCB resistance. To test the hypothesis, we used Hsp27 inhibitor, quercetin, and Hsp27 siRNA to treat glioblastoma cells. Our results showed that, with the pretreatment of quercetin or Hsp27 siRNA, Atg7 expression was markedly decreased in U87 or U251 cells, even in t-AUCB treated cells (Fig. 2a). Contrarily, knockdown of Atg7 expression by siRNA didn’t reduce Hsp27 expression (Fig. 2b).
Atg7 is downstream of Hsp27 and participates the resistance of glioblastoma cells to t-AUCB. a U87 cells were treated with DMSO, 200 μM t-AUCB, 30 μM quercetin or 30 μM quercetin plus 200 μM t-AUCB; U251 cells were treated with DMSO, 200 μM t-AUCB, Hsp27 siRNA or Hsp27 siRNA plus 200 μM t-AUCB. Western blotting was performed to test Hsp27 and Atg7 expression. β-actin served as loading control. b U87 cells were treated with DMSO, 200 μM t-AUCB, Atg7 siRNA or Atg7 siRNA plus 200 μM t-AUCB. Western blotting was performed to test Hsp27 and Atg7 expression. β-actin served as loading control. c U87 and U251 cells were respectively treated with DMSO (vehicle control), Lipofectamine (negative congtrol), Negative control siRNA, 200 μM t-AUCB, Hsp27 siRNA, Atg7 siRNA, Hsp27 siRNA plus 200 μM t-AUCB or Atg7 siRNA plus 200 μM t-AUCB. Cell growth was tested using CCK-8 kit. Compare with DMSO, #P > 0.05; Compare with DMSO, *P < 0.05; Compare with t-AUCB, **P < 0.05; Compare with siRNA, ***P < 0.05
To investigate whether Atg7 participates in resistance to t-AUCB, U87 and U251 cells were respectively treated with DMSO (vehicle control), Lipofectamine (transfection reagent), Negative control siRNA, 200 μM t-AUCB, Hsp27 siRNA, Atg7 siRNA, Hsp27 siRNA plus 200 μM t-AUCB or Atg7 siRNA plus 200 μM t-AUCB for 48 h. Cell growth was tested using CCK-8 kit. As the results shown in Fig. 2c, with the downregulation of Hsp27 or Atg7 by siRNA, t-AUCB induces more intense cell growth inhibition (P < 0.05). Cells with knockdown of Hsp27 are more sensitive to t-AUCB than those with knockdown of Atg7 (P < 0.05). Since Atg7 is downstream of Hsp27 in t-AUCB treated glioblastoma cells, it suggests that Atg7 may not be the only factor that participates in Hsp27-induced cell resistance to t-AUCB.
Quercetin strengthens t-AUCB-induced cell growth inhibition by autophagy blockage
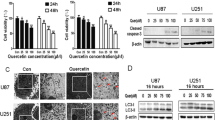
Quercetin inhibits Atg7 expression by Hsp27 inhibition. To investigate whether quercetin strengthens t-AUCB-induced cell growth inhibition, U87 and U251 cells were treated with DMSO (vehicle control), 200 μM t-AUCB, 5, 15, 30 μM quercetin or 200 μM t-AUCB plus 15 or 30 μM quercetin for 48 h. Cell growth was tested using CCK-8 kit. Our results showed that, quercetin induces cell growth inhibition in a concentration-depended manner since 15 μM, and strengthens t-AUCB induced cell growth inhibition (Fig. 3a).
Quercetin strengthens t-AUCB-induced cell growth inhibition by autophagy blockage. a U87 and U251 cells were treated with DMSO (vehicle control), 200 μM t-AUCB, 5, 15, 30 μM quercetin or 200 μM t-AUCB plus 15 or 30 μM quercetin for 48 h. Cell growth was tested using CCK-8 kit. Compare with DMSO, #P > 0.05; Compare with DMSO, *P < 0.05; Compare with t-AUCB, **P < 0.05; Compare with quercetin, ***P < 0.05 b U87 cells were treated with DMSO (vehicle control), 200 μM t-AUCB, 30 μM quercetin or 30 μM quercetin plus 200 μM t-AUCB for 24 h, and then analyzed by transmission electron microscopy. Black arrows indicate the autophagic vacuoles. c The computation of the autophagic vacuoles per cell (*P < 0.05)
We hypothesized that quercetin may strengthens t-AUCB-induced cell growth inhibition by blocking t-AUCB-induced autophagy. To test this hypothesis, U87 cells were treated with DMSO (vehicle control), 200 μM t-AUCB, 30 μM quercetin or 30 μM quercetin plus 200 μM t-AUCB for 24 h, and then analyzed by transmission electron microscopy. Membrane-bound vacuoles were counted as autophagosomes. The results in Fig. 3b, c showed that quercetin blocks t-AUCB-induced autophagy.
Discussion
Since autophagy is activated during a time of cell stress and serves as an escape mechanism for cells exposed to cytotoxic agents, more and more studies unveil the roles of autophagy in chemoresistance during cancer treatment [16–19]. However, it is complex to enhance the efficacy of anticancer therapy by modulating autophagy. The role of anticancer therapy-induced autophagy in cells is tumor-type or stage dependent. Either suppression of cell-survival autophagy or enhancement of cytotoxic autophagy may strengthen anticancer effects [20]. Thus, appropriate modification of autophagy should be carefully concerned. Autophagy has also been demonstrated to participate in chemo- or radioresistance to glioma [12, 21]. Plenty of findings showed that inhibition of autophagy might be a potential strategy to strengthen cytotoxic effect of chemo- or radiotherapy to glioma [11, 12, 22–25]. However, a recent phase I/II trial of hydroxychloroquine (HCQ) in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme declared that, autophagy inhibition was not consistently achieved in patients treated with HCQ in maximum tolerated dose, and no significant improvement in overall survival was observed, although their preclinical studies indicated autophagy inhibition with HCQ can augment the efficacy of DNA-damaging therapy [26]. Thus, it is necessary to test or develop lower-toxicity compounds that can achieve more consistent inhibition of autophagy.
In the present study, we found autophagy participates in acquired resistance of glioblastoma cells to t-AUCB as expected. We previously demonstrated that Hsp27 confers resistance of glioblastoma cells to t-AUCB, and t-AUCB induces significant apoptosis after blocking the activation of Hsp27 [2]. Others reported that Atg7 is downstream of Hsp27 [15]. Thus, we hypothesized that Atg7 may be regulated by Hsp27 and involved in autophagy modulation and t-AUCB resistance. The gene ATG7 (autophagy related 7) encodes an E1-like activating enzyme that is essential for autophagy and cytoplasmic to vacuole transport [27]. Atg7 functions as an enzyme necessary for Atg12-Atg5 conjugation and regulates the LC3 transforming from LC3-I to LC3-II, which is essential for autophagosome formation [14]. In Drosophila, Atg7-null mutants are short-lived and hypersensitive to starvation and oxidative stress because of blockage of autophagy [28]. Atg7 knockout mice exhibited neurodegeneration because of suppression of basal autophagy in the central nervous system [29]. Herein, our data showed autophagy induction and Atg7 overexpression in t-AUCB treated glioblastoma cells, and demonstrated that RNA interference-mediated knockdown of Atg7 sensitizes glioblastoma cells to t-AUCB and Atg7 inhibition by quercetin partially recovered t-AUCB-induced autophagy, indicating Atg7 participate t-AUCB resistance via autophagy modulation.
To investigate whether Hsp27 regulates Atg7 expression, we treated glioblastoma cells with quercetin, a Hsp27 inhibitor, which significantly inhibits expression of both Hsp27 and Atg7. Then, we found Hsp27 knockdown by RNA-interference suppresses Atg7 expression. But Atg7 knockdown do not affect Hsp27 expression. These data suggested that Atg7 acts downstream of Hsp27 in the regulation of autophagy and participates in conferring t-AUCB resistance. Moreover, we found Hsp27 silencing showed better effect than Atg7, indicating that Atg7 may only one of the factors that participate in Hsp27-induced cell resistance to t-AUCB. As a Hsp27 inhibitor, quercetin suppresses Atg7 expression quite efficiently (Fig. 2a). Atg7 deficiency caused by quercetin recovered t-AUCB-induced autophagy and augmented the cytotoxic effect of t-AUCB. Moreover, quercetin strengthened t-AUCB-induced cell death more efficiently than Atg7 or Hsp27 knockdown did, suggesting that quercetin may also function by other ways except Hsp27 and Atg7 inhibition.
Quercetin is a bioflavonoid distributed in plants [30]. Its antitumor effect has been displayed in several cancer cells [31]. Quercetin is low-toxicity and has been studied in clinical trials [32], which make it to be a possible replacement to hydroxychloroquine in autophagy inhibition for glioma treatment. Our data demonstrated for the first time that quercetin recovers t-AUCB-induced autophagy and eliminates the resistance to t-AUCB in glioblastoma cells by Hsp27 and Atg7 inhibition.
In conclusion, our data demonstrated that t-AUCB induces autophagy in glioblastoma cells and regulates multiple autophagy related-gene expression. t-AUCB induces overexpression of Atg7 which is downstream of Hsp27 and participates the resistance of glioblastoma cells to t-AUCB treatment. Hsp27 inhibitor quercetin suppresses Atg7 expression and strengthens t-AUCB-induced cell death by autophagy blockage. Combination of quercetin and t-AUCB may be a potential strategy for glioblastoma treatment.
References
Li J, Liu H, Xing B, Yu Y, Wang H, Chen G, Gu B, Zhang G, Wei D, Gu P, Li M, Hu W (2012) t-AUCB, an improved sEH inhibitor, suppresses human glioblastoma cell growth by activating NF-kappaB-p65. J Neurooncol 108:385–393. doi:10.1007/s11060-012-0841-4
Li J, Hu W, Lan Q (2012) The apoptosis-resistance in t-AUCB-treated glioblastoma cells depends on activation of Hsp27. J Neurooncol 110:187–194. doi:10.1007/s11060-012-0963-8
Li JY, Li RJ, Wang HD (2014) Gamma-secretase inhibitor DAPT sensitizes t-AUCB-induced apoptosis of human glioblastoma cells in vitro via blocking the p38 MAPK/MAPKAPK2/Hsp27 pathway. Acta Pharmacol Sin 35:825–831. doi:10.1038/aps.2013.195
He M, Luo M, Liu Q, Chen J, Li K, Zheng M, Weng Y, Ouyang L, Liu A (2016) Combination treatment with fasudil and clioquinol produces synergistic anti-tumor effects in U87 glioblastoma cells by activating apoptosis and autophagy. J Neurooncol. doi:10.1007/s11060-015-2044-2
Hori YS, Hosoda R, Akiyama Y, Sebori R, Wanibuchi M, Mikami T, Sugino T, Suzuki K, Maruyama M, Tsukamoto M, Mikuni N, Horio Y, Kuno A (2015) Chloroquine potentiates temozolomide cytotoxicity by inhibiting mitochondrial autophagy in glioma cells. J Neurooncol 122:11–20. doi:10.1007/s11060-014-1686-9
Levy JM, Thorburn A (2012) Modulation of pediatric brain tumor autophagy and chemosensitivity. J Neurooncol 106:281–290. doi:10.1007/s11060-011-0684-4
Bo Q, Ma S, Han Q, Wang FE, Li X, Zhang Y (2015) Role of autophagy in photoreceptor cell survival and death. Crit Rev Eukaryot Gene Expr 25:23–32
Nah J, Yuan J, Jung YK (2015) Autophagy in neurodegenerative diseases: from mechanism to therapeutic approach. Mol Cells 38:381–389. doi:10.14348/molcells.2015.0034
Jardon MA, Rothe K, Bortnik S, Vezenkov L, Jiang X, Young RN, Lum JJ, Gorski SM (2013) Autophagy: from structure to metabolism to therapeutic regulation. Autophagy 9:2180–2182. doi:10.4161/auto.26378
Wu T, Wang MC, Jing L, Liu ZY, Guo H, Liu Y, Bai YY, Cheng YZ, Nan KJ, Liang X (2015) Autophagy facilitates lung adenocarcinoma resistance to cisplatin treatment by activation of AMPK/mTOR signaling pathway. Drug Des Devel Ther 9:6421–6431. doi:10.2147/DDDT.S95606
Shteingauz A, Boyango I, Naroditsky I, Hammond E, Gruber M, Doweck I, Ilan N, Vlodavsky I (2015) Heparanase enhances tumor growth and chemoresistance by promoting autophagy. Cancer Res 75:3946–3957. doi:10.1158/0008-5472.CAN-15-0037
Lomonaco SL, Finniss S, Xiang C, Decarvalho A, Umansky F, Kalkanis SN, Mikkelsen T, Brodie C (2009) The induction of autophagy by gamma-radiation contributes to the radioresistance of glioma stem cells. Int J Cancer 125:717–722. doi:10.1002/ijc.24402
Liu JY, Tsai HJ, Hwang SH, Jones PD, Morisseau C, Hammock BD (2009) Pharmacokinetic optimization of four soluble epoxide hydrolase inhibitors for use in a murine model of inflammation. Br J Pharmacol 156:284–296. doi:10.1111/j.1476-5381.2008.00009.x
Mandelbaum J, Rollins N, Shah P, Bowman D, Lee JY, Tayber O, Bernard H, LeRoy P, Li P, Koenig E, Brownell JE, D’Amore N (2015) Identification of a lung cancer cell line deficient in atg7-dependent autophagy. Autophagy. doi:10.1080/15548627.2015.1056966
Chen SF, Kang ML, Chen YC, Tang HW, Huang CW, Li WH, Lin CP, Wang CY, Wang PY, Chen GC, Wang HD (2012) Autophagy-related gene 7 is downstream of heat shock protein 27 in the regulation of eye morphology, polyglutamine toxicity, and lifespan in Drosophila. J Biomed Sci 19:52. doi:10.1186/1423-0127-19-52
Tan Q, Wang H, Hu Y, Hu M, Li X, Aodengqimuge Ma Y, Wei C, Song L (2015) Src/STAT3-dependent heme oxygenase-1 induction mediates chemoresistance of breast cancer cells to doxorubicin by promoting autophagy. Cancer Sci 106:1023–1032. doi:10.1111/cas.12712
Yang HZ, Ma Y, Zhou Y, Xu LM, Chen XJ, Ding WB, Zou HB (2015) Autophagy contributes to the enrichment and survival of colorectal cancer stem cells under oxaliplatin treatment. Cancer Lett 361:128–136. doi:10.1016/j.canlet.2015.02.045
Liu Y, Zhao L, Ju Y, Li W, Zhang M, Jiao Y, Zhang J, Wang S, Wang Y, Zhao M, Zhang B, Zhao Y (2014) A novel androstenedione derivative induces ROS-mediated autophagy and attenuates drug resistance in osteosarcoma by inhibiting macrophage migration inhibitory factor (MIF). Cell Death Dis 5:e1361. doi:10.1038/cddis.2014.300
Chittaranjan S, Bortnik S, Dragowska WH, Xu J, Abeysundara N, Leung A, Go NE, DeVorkin L, Weppler SA, Gelmon K, Yapp DT, Bally MB, Gorski SM (2014) Autophagy inhibition augments the anticancer effects of epirubicin treatment in anthracycline-sensitive and -resistant triple-negative breast cancer. Clin Cancer Res 20:3159–3173. doi:10.1158/1078-0432.CCR-13-2060
Shingu T, Fujiwara K, Bogler O, Akiyama Y, Moritake K, Shinojima N, Tamada Y, Yokoyama T, Kondo S (2009) Stage-specific effect of inhibition of autophagy on chemotherapy-induced cytotoxicity. Autophagy 5:537–539
Zou Y, Wang Q, Li B, Xie B, Wang W (2014) Temozolomide induces autophagy via ATMAMPKULK1 pathways in glioma. Mol Med Rep 10:411–416. doi:10.3892/mmr.2014.2151
Pratt J, Annabi B (2014) Induction of autophagy biomarker BNIP3 requires a JAK2/STAT3 and MT1-MMP signaling interplay in Concanavalin-A-activated U87 glioblastoma cells. Cell Signal 26:917–924. doi:10.1016/j.cellsig.2014.01.012
Chen Y, Meng D, Wang H, Sun R, Wang D, Wang S, Fan J, Zhao Y, Wang J, Yang S, Huai C, Song X, Qin R, Xu T, Yun D, Hu L, Yang J, Zhang X, Chen H, Chen J, Chen H, Lu D (2015) VAMP8 facilitates cellular proliferation and temozolomide resistance in human glioma cells. Neurooncology 17:407–418. doi:10.1093/neuonc/nou219
Nabissi M, Morelli MB, Amantini C, Liberati S, Santoni M, Ricci-Vitiani L, Pallini R, Santoni G (2015) Cannabidiol stimulates Aml-1a-dependent glial differentiation and inhibits glioma stem-like cells proliferation by inducing autophagy in a TRPV2-dependent manner. Int J Cancer 137:1855–1869. doi:10.1002/ijc.29573
Palumbo S, Tini P, Toscano M, Allavena G, Angeletti F, Manai F, Miracco C, Comincini S, Pirtoli L (2014) Combined EGFR and autophagy modulation impairs cell migration and enhances radiosensitivity in human glioblastoma cells. J Cell Physiol 229:1863–1873. doi:10.1002/jcp.24640
Rosenfeld MR, Ye X, Supko JG, Desideri S, Grossman SA, Brem S, Mikkelson T, Wang D, Chang YC, Hu J, McAfee Q, Fisher J, Troxel AB, Piao S, Heitjan DF, Tan KS, Pontiggia L, O’Dwyer PJ, Davis LE, Amaravadi RK (2014) A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy 10:1359–1368. doi:10.4161/auto.28984
He C, Klionsky DJ (2009) Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43:67–93. doi:10.1146/annurev-genet-102808-114910
Juhasz G, Erdi B, Sass M, Neufeld TP (2007) Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev 21:3061–3066. doi:10.1101/gad.1600707
Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441:880–884. doi:10.1038/nature04723
Wang G, Wang JJ, Chen XL, Du SM, Li DS, Pei ZJ, Lan H, Wu LB (2013) The JAK2/STAT3 and mitochondrial pathways are essential for quercetin nanoliposome-induced C6 glioma cell death. Cell Death Dis 4:e746. doi:10.1038/cddis.2013.242
Hsu HS, Lin JH, Huang WC, Hsu TW, Su K, Chiou SH, Tsai YT, Hung SC (2011) Chemoresistance of lung cancer stemlike cells depends on activation of Hsp27. Cancer 117:1516–1528. doi:10.1002/cncr.25599
Shoskes DA, Zeitlin SI, Shahed A, Rajfer J (1999) Quercetin in men with category III chronic prostatitis: a preliminary prospective, double-blind, placebo-controlled trial. Urology 54:960–963
Acknowledgments
We thank Professor Bruce D. Hammock for providing the sEH inhibitor t-AUCB. This study was supported by research fund from National Natural Science Foundation of China (No. 81301905).
Conflict of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Junyang Li and Chao Tang have contributed equally to this work.
Chao Tang—co-first author.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, J., Tang, C., Li, L. et al. Quercetin blocks t-AUCB-induced autophagy by Hsp27 and Atg7 inhibition in glioblastoma cells in vitro. J Neurooncol 129, 39–45 (2016). https://doi.org/10.1007/s11060-016-2149-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-016-2149-2