Abstract
Insulin like growth factor binding protein 4 (IGFBP4) regulates growth and development of tissues and organs by negatively regulating IGF signaling. Among most cancers, IGFBP4 has growth inhibitory role and reported as a down-regulated gene, except for renal cell carcinoma, wherein IGFBP4 promotes tumor progression. IGFBP4 expression has been shown to be higher in increasing grades of astrocytoma. However, the functional role of IGFBP4 in gliomas has not been explored. Surgical biopsies of 20 normal brain and 198 astrocytoma samples were analyzed for IGFBP4 expression by qRT-PCR. Highest expression of IGFBP4 mRNA was seen in GBM tumors compared to control brain tissues (median log2 of 2.035, p < 0.0001). Immunohistochemical analysis of 53 tissue samples revealed predominant nuclear staining of IGFBP4, seen maximally in GBMs when compared to DA and AA tumors (median LI = 29.12 ± 16.943, p < 0.001). Over expression of IGFBP4 in U343 glioma cells resulted in up-regulation of molecules involved in tumor growth, EMT and invasion such as pAkt, pErk, Vimentin, and N-cadherin and down-regulation of E-cadherin. Functionally, IGFBP4 over expression in these cells resulted in increased proliferation, migration and invasion as assessed by MTT, transwell migration, and Matrigel invasion assays. These findings were confirmed upon IGFBP4 knockdown in U251 glioma cells. Our data suggest a pro-tumorigenic role for IGFBP4 in glioma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The insulin-like growth factor (IGF) signaling pathway is implicated in the development and progression of many different malignancies [1]. Several studies have documented the role of IGF’s, their cognate receptors and binding proteins in the progression of human neoplasia [2–6]. The IGF binding proteins (IGFBP) bind IGF-I and IGF-II, prolongs their half-life in serum and thereby regulate IGF actions [7, 8]. Increased expression of IGFBPs 2, 3, 5 and 7 has been reported in astrocytomas and correlate with poor outcome in GBM patients [9–11]. IGFBP2, 5 and 7 have been shown to be involved in glioma progression [5, 10, 12].
Insulin-like growth factor binding protein 4 (IGFBP4), a 34 kDa protein and one among six high affinity IGFBP’s binds to both IGF1 and 2 [13]. IGFBP4 is reported to regulate the biological actions of IGF1 and 2 in vivo in a positive or negative manner [14]. IGFs may regulate their own availability through proteolytic degradation of IGFBP4 [15]. IGFBP4 expression was found to be lower in tumor cells of several neoplasms such as neuroblastoma [16], lung [17, 18], breast [19], colon [20], thyroid [21], compared to respective normal tissues. In breast cancer patients, IGFBP4 expression was considered as an independent prognostic factor with better prognosis in patients with ER (+) breast tumors [22]. Over expression of IGFBP4 has been shown to inhibit the growth of colon and prostate cancer cells in vivo, [23, 24] suggesting a tumor suppressor role for IGFBP4. However, IGFBP4 has been shown to have tumor promoting effects in renal cell carcinoma as determined by in vitro and in vivo studies [25]. IGFBP4 expression pattern has been shown to be progressively elevated across diffused astrocytoma (DA) to glioblastoma (GBM) suggesting a pro-tumorigenic role for IGFBP4 in progressive astrocytoma [26]. However, a direct functional role of IGFBP4 in glioma tumorigenesis has not been reported.
In this study we provide data on the expression pattern of IGFBP4 in different grades of astrocytoma and by over expression and knock down experiments we demonstrate a role for IGFBP4 in proliferation, promotion of EMT, migration and invasion of GBM cells.
Materials and methods
Patient and tissue samples
Diffusely infiltrating astrocytoma tissues of different grades (DA, AA and GBM) and control tissues were obtained by surgical resection performed on patients at the National Institute of Mental Health and Neurosciences and Sri Satya Sai Institute of Higher Medical Sciences, Bangalore, India, as previously described [9]. This study has been approved by the respective institutional review boards and informed consent was obtained from each patient. Tissues were bisected; a portion was minced, placed in RNA later (Ambion Inc., USA) and stored at –70o C for RNA isolation. The other portion was fixed in 10 % buffered neutral formalin, processed for paraffin sections and used for histopathology and IHC.
Cell culture and transfection
Human GBM cell lines U343, U251 were kind gift from K. Somasundaram, IISc Bangalore. Both cell lines were cultured in DMEM (Dulbecco’s modified Eagle’s medium, Sigma-Aldrich) supplemented with 3.7 g/l sodium bicarbonate (Sigma-Aldrich, USA) and 10 % (v/v) FBS (Fetal bovine serum, Invitrogen) and 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco) at 37 °C in a 5 % CO2 atmosphere.
IGFBP4 expression plasmid (pCMV6-AC-IGFBP4) (cat no: sc319642) was obtained from OriGene Technologies (Rockville, MD). Transient transfection of plasmid DNA into cells was performed using lipofectamine 2000 (Invitrogen, USA) according to manufacturer’s instructions.
Knockdown of IGFBP4 by siRNA in U251 cell line
U251 cells were transfected with 100 nM siRNA against IGFBP4 (cat no: M-012583-01-0010) (Dharmacon RNAi Technologies, Thermo scientific) or a scrambled siRNA (cat no: D-001206-14-20) (Dharmacon RNAi Technologies, Thermo scientific), for 96 h. Transfections were performed using Oligofectamine reagent (Invitrogen, India) according to manufacturer’s instructions. The efficiency of knockdown was assessed by western blot analysis.
RNA extraction and RT-PCR
Total RNA isolation, cDNA synthesis and semi quantitative RT-PCR was performed as described earlier [27]. Real-time quantitative PCR was carried out in the ABI PRISM 7900HT sequence detection system (Applied Biosystems, USA). A total of 218 tissues (16 DA, 50 AA, 132 GBM and 20 control brain tissues) were processed for qPCR using the DyNAmo™ HS SYBR Green qPCR kit (Finnzymes, Finland). For normalization, the mean expression levels of ribosomal protein L35A (RP-L35A), 1-acylglycerol- 3-phosphate O-acyltransferase 1 (AGPAT1), ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9; ATP5G1), and glycyl-tRNA synthetase (GARS) were used and the data analysis was performed as described earlier [28]. Normal brain tissue samples (n = 20) from epilepsy patients were used as reference. Primer sequence of the genes are listed in supplementary, Table S1.
Western blot analysis
For protein extraction, cells were lysed in RIPA buffer as described earlier [29]. For each sample, 100 μg total proteins were separated on SDS-PAGE gels before transferring to PVDF membrane (Immobilin P, Millipore). Membranes were then incubated with the appropriate primary antibody (IGFBP4, Santa Cruz, USA; B-actin, Sigma, USA; IGF1R, p-IGF1R Vimentin, N-cadherin, E-cadherin, p-Akt, Total Akt, p-Erk and total Erk, Cell signaling technology, USA) and detection was performed as recommended by the manufacturers with HRP-conjugated secondary antibody using the Pearson Western blotting detection system (Thermo scientific).
Immunocytochemistry
For immunocytochemistry, 20,000 cells were plated on sterile glass cover slip. Transfection was performed as described before and after recovery, cells were washed with DPBS and fixed with methanol at −20 °C and blocked with FBS (10 %) in DPBS buffer for 1 h. IGFBP4 primary antibody (1:50 dilution) was added and incubated overnight with gentle rocking. After washing, secondary antibody (Alexa flour 488; dilution factor of 1:1,500 in antibody buffer with 2 % sera) was added and incubated for 1 h. Cover slips were washed with DPBS, counter stained for nuclei using 0.33 mg/ml solution of Propidium iodide (Sigma Aldrich, USA), mounted on glass slide with antifade solution (Prolong gold, Invitrogen Inc, USA) and image was captured on Zeiss Confocal Microscope (LSM 510 Meta).
Cell proliferation assay
Cell proliferation was estimated by MTT assay [30]. Briefly, equal number (5,000) of cells were plated in triplicate in 96 well tissue culture plates and left to grow for 48 h (experimental plate). 48 h after cell plating, 20 μl of 5 mg/ml MTT (Sigma Aldrich, USA) was added to the cells to terminate the assay. 3 h after MTT addition, the medium was removed from the wells and 200 μl of DMSO was added to the wells and absorbance was measured at 595 nm.
Immunohistochemistry
Paraffin sections (4 μm) from the tumor tissue and control samples were collected on silane-coated slides and immunohistochemistry for the protein expression of IGFBP4, was carried out on 53 samples that included 4 control brain tissues, 9 DA, 10 AA, and 30 GBM tumors. The IGFBP4 antibody was from Santa Cruz Biotechnology and pAkt (ser 473) was from cell signaling technologies (CST, USA). Processing of tissue sections for immunochemistry and assessment were performed as described before [9]. A visual semi-quantitative grading scale was applied to assess the intensity of the immunoreactivity as follows: zero (0) if the staining was absent, 1+ if it was mild and 2+ if it was strong. Only 2+ staining intensity was considered for analysis. The IGFBP4 and pAkt labeling index (LI) was expressed as a percentage of cells that showed 2+ positive staining among the total number of cells that were counted.
Transwell migration assay
The BD BioCoat Matrigel Cell Culture control Inserts (BD Biosciences, USA) were used for migration assays. Briefly, the control inserts were filled with cell suspension after transfection and recovery (5 × 104 cells in 0.5 ml of DMEM), and placed in a well of a 24-well plate. At 22 h after incubation the cells on the upper side of the filters were removed with cotton-tipped swabs. Migrated cells on the lower side of the filters were fixed and stained with 0.05 % Crystal Violet [31]. The cells on the underside of the filters were viewed under a microscope and counted.
Matrigel invasion assay
The BD BioCoat Matrigel Cell Culture Inserts (BD Biosciences, USA) were used for invasion assays. Briefly, the control inserts or matrigel-coated inserts were rehydrated with plain DMEM for 2 h before use. These were then filled with cells after transfection and recovery (5 × 104 cells in 0.5 ml of DMEM), and placed in a well of a 24-well plate. At 22 h after incubation the cells on the upper side of the filters were removed with cotton-tipped swabs. Migrated and/or invaded cells on the lower side of the filters were fixed and stained with 0.05 % Crystal Violet. The cells on the underside of the filters were viewed under a microscope and counted.
Statistical analysis
Significance of differential expression of IGFBP4 and pAkt was tested by a non-parametric test, Kruskal–Wallis One-Way Analysis of Variance on Ranks, followed by Dunn’s post hoc test. Statistical analyses for all semi quantitative RT-PCR analysis (n = 3) were performed with GraphPad Prism 5.0 Software (GraphPad Software, San Diego).
Results
IGFBP4 expression in glioma tumors
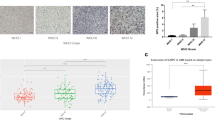
We analyzed the expression pattern of IGFBP4 across different grades of 198 diffusely infiltrating astrocytoma that included 16 DA, 50 AA, 132 GBM tumors and 20 control brain tissues using the SYBRGreen based qPCR assay, as described in the methods section. Compared to the expression in control brain tissues, IGFBP4 showed up-regulation in 85 % (118/132, median log2 value 2.035) of GBM tumors (p < 0.0001), while it was up-regulated only in 46 % (23/50, median log2 ratio of 0.25) of AA (p = 0.095), which is not significant. In DA tumors, only 25 % (4/10, median log2 ratio of −0.45) showed up-regulation, which is also not significant (p = 0.1626) (Fig. 1a). Taken together, IGFBP4 expression was up-regulated in GBMs as compared to control tissues. In order to confirm the expression of IGFBP4 protein in glioma tissues, IHC was performed on tissue sections of grades of astrocytoma. Highest intensity of IGFBP4 staining was observed in GBM, compared to control and other grades of astrocytoma (Fig. 1b). Interestingly IGFBP4 showed nuclear localization. In the normal brain, faint nuclear staining (1+) was noted in most of the neurons, but not in astrocytes and oligodendroglial cells. A significant differential protein expression of IGFBP4 was observed in diffusely infiltrating astrocytomas compared to normal tissues (Fig. 1b). The mean ± SD LI for IGFBP4 in GBM was 29.12 ± 16.943, while in AA, DA and control it was 9.00 ± 13.784, 16.54 ± 16.631 and 0.00 ± 0.000, respectively. The comparison between groups by Kruskal–Wallis One way ANOVA followed by Dunn’s post hoc test showed that the difference between GBM and control; GBM and DA; GBM and AA was significant (p < 0.0001).
Expression of IGFBP4 in diffusely infiltrating astrocytoma. a Relative mRNA expression of IGFBP4 in various grades of 198 diffusely infiltrating astrocytomas and 20 control brain tissues are shown as scatter plots. The log 2-transformed gene expression ratios obtained from qPCR analyses of the tumor tissues against control tissues are plotted. A significant difference in the median log 2 ratio of IGFBP4 was observed between GBM, AA, DA, and control tissues by Kruskal–Wallis one-way ANOVA followed by Tukey’s post hoc test (p < 0.05). Bars show group medians. b Representative micrographs showing IHC staining pattern of IGFBP4 in 9 DA, 10 AA and 30 GBM tissues. Increasing IGFBP4 expression was seen with increasing grades compared to control brain tissues. IGFBP4 protein shows predominant nuclear labeling (arrows). All original magnifications are ×20 for IGFBP4
IGFBP4 influences the proliferation of glioma cells
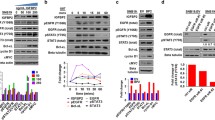
In order to understand the role of IGFBP4 on in vitro proliferation of glioma cells, IGFBP4 was transiently over expressed in U343 glioma cells. The increased expression was confirmed by RT-PCR (Fig. 2a), western blot (Fig. 2b) and immunocytochemistry (Fig. S1A). IGFBP4 over expression resulted in increased proliferation as determined by MTT assay (Fig. 2c). IGFBP4’s role in proliferation was confirmed by increased phosphorylation status of Erk and Akt in IGFBP4 transfected U343 cells when compared with vector transfected cells (Fig. 2b). In good correlation IGFBP4 knockdown (Figs. 2d, e and S1b) resulted in decreased proliferation (Fig. 2f) and also down regulation of pAkt and pErk (Fig. 2e) in U251 cells. Intriguingly, IGFBP4 protein localized to cytoplasm in U343 cells and is nuclear localized in U251 cells suggesting that the IGFBP4 localization is cell type specific (Fig. S1b).
IGFBP 4 promotes proliferation of glioma cells. a Quantitation of IGFBP4 RNA upon over expression in U343 cells, normalized with HPRT expression (n = 3). b Representative immunoblots showing protein expression pattern of IGFBP4, pAkt, pErk and c MTT assay to determine proliferation of IGFBP4 over expressing cells compared to vector transfected cells. d IGFBP4 expression upon knock down in U251 cells using siRNA normalized to expression of RPL35A (n = 3); e Protein expression pattern and f MTT assay showing proliferation pattern. Statistical significance is depicted over the respective bars. The gel images are shown in supplementary Fig. S2 a&b
In order to determine if the regulation of pAkt by IGFBP4 in cell lines reflects their expression status in tumors, IHC for pAkt was performed on astrocytomas that were previously stained for IGFBP4 expression. As shown in Fig. 3, IGFBP4 expression by IHC correlated with pAkt expression in all grades of diffusely infiltrating astrocytomas. pAkt expression was predominantly cytoplasmic. A significant differential protein expression of pAkt was also observed in diffusely infiltrating astrocytomas (p < 0.001).
p-Akt expression correlates with IGFBP4 expression in different grades of astrocytoma a Representative micrographs showing immunohistochemical staining pattern of p-Akt (arrows) performed as described in the methods in DA (9), AA (10) and GBM (30) tissue sections. b Scatter diagram depicting labeling index of each tissue sample. All original magnifications are ×20
IGFBP4 expression induces EMT, migration and invasion of glioma cells
Transient over expression of IGFBP4 in U343 cells resulted in the regulation of EMT markers such as down-regulation of E-cadherin, and up-regulation of vimentin, N-cadherin, snail, slug, and twist (Figs. 4a, S2A). These results were also confirmed by immunoblot analysis (Fig. 4b). Since cells undergoing EMT acquire an invasive phenotype, we analysed effect of IGFBP4 over expression on the migration and invasion potential of U343 cells. In the transwell matrigel invasion assay, U343 cells over expressing IGFBP4 showed increased invasion (Fig. 4d), compared with vector pCMV6 transfected cells though there was no significant effect on migration of these cells (Fig. 4c).
Effect of IGFBP4 over expression on EMT, migration and invasion of glioma cells. a Represents fold change ratio of EMT genes upon IGFBP4 overexpression in U343 cell line compared to vector control as determined by semi-quantitative RT‐PCR analysis, normalized with HPRT gene (n = 3). b Expression of EMT markers in U343 cells transfected with empty plasmid (vector) or IGFBP4 expressing plasmid. Over expression of IGFBP4 was assessed using polyclonal antibody against human IGFBP4 by western blot analysis. β-actin served as a loading control. c Graph showing migration of IGFBP4 over expressing U343 cells as compared to vector control. d Invasion assay of IGFBP4 over expressing U343 cells. 5 × 104 cells were placed in each chamber at the beginning of the assay. The number of invaded cells was significantly increased in IGFBP4 over expressing U343 cells as compared with vector control (p = < 0.001). Each experiment was performed in triplicates
These results were also confirmed by IGFBP4 knockdown in U251 cells using siRNA which resulted in the down-regulation of mRNAs of EMT markers Twist and N-cadherin and up-regulation of E-cadherin (Figs. 5a, S2B). There was no detectable mRNA for snail and slug in these cells (data not shown). Immunoblot analysis of lysates prepared from IGFBP4 knock down U251 cells confirmed the RNA data (Fig. 5b) except for Vimentin. There was no detectable change in the Vimentin RNA in IGFBP4 knock down cells whereas protein showed a decrease. The reasons for this discrepancy are not known, at the moment. Knock down of IGFBP4 expression by siRNA in U251 cells resulted in decreased migration (Fig. 5c) and invasion (Fig. 5d). Taken together, these data suggest that IGFBP4 induces EMT, migration and invasion in glioma cells.
Effect of IGFBP4 knockdown on EMT, migration and invasion of glioma cells. a Represents fold change ratio of EMT genes upon IGFBP4 knockdown in U251 cell line compared to vector control as determined by semi-quantitative RT‐PCR analysis, normalized with RPL35a gene (n = 3). b Expression of EMT markers in U251 cells transfected with IGFBP4 si-RNA or scrambled si-RNA. Knockdown of IGFBP4 was assessed using polyclonal antibody against human IGFBP4 by western blot analysis. β-actin served as a loading control. c Graph showing decreased migration of U251 cells with IGFBP4 knockdown as compared to scrambled siRNA transfected cells. d Invasion assay performed on IGFBP4 knock down U251 cells. 2 × 104 cells were placed in each chamber at the beginning of the assay. The number of invaded cells was significantly decreased in IGFBP4 siRNA transfected U251ncells as compared with Scrambled siRNA transfected cells (p = < 0.01). Each experiment was performed in triplicates
Discussion
Primarily IGFBP4 is known to exhibit growth inhibitory effects on cells which are dependent on IGFs for growth and survival [32–34] except in metastatic renal cell carcinoma where IGFBP4 was shown to have tumor promoting effects [25]. In contrast to most reports suggesting a tumor suppressing role for IGFBP4 in many cancers, our data revealed a tumor promoting role in GBM. IHC data from glioma patient samples revealed increased expression of IGFBP4 in different grades of diffusely infiltrating astrocytoma with maximal expression in GBM. Notably, unlike other IGFBP isoforms, IGFBP4 showed a clear nuclear labeling by IHC in these tumors. IGFBP4 nuclear localization has also been reported in breast cancer and normal breast tissues [8]. In order to determine the functional significance of IGFBP4, we performed over expression and knock down studies in U343 and U251 glioma cells, respectively. Our data suggest tumor promoting roles for IGFBP4. IGFBP4 influences proliferation of glioma cells and interestingly, IGFBP4 positively regulates the phosphorylation status of Erk and Akt in glioma cells. This finding is in contrast to the reported actions of IGFBP4 inhibiting IGF actions by affecting Erk and Akt pathways in bone, rat neuronal and colon cancer cell [35], [36–38]. Activation of Erk/Akt phosphorylation by IGFBP4 shown in this study is similar to the actions of IGFBP2 reported in glioma cells [39, 40]. However the mechanism of activation of Erk/Akt by IGFBP4 is not known. We also present data on the increased pAkt in GBMs by IHC. Since, multiple pathways are known to regulate Erk/Akt pathways; it is not clear whether Erk/Akt regulation by IGFBP4 requires cross talk with other signaling pathways. Our study revealed regulation of EMT markers such as vimentin, N-cadherin, E-cadherin, and transcription factors such as snail, slug and twist by IGFBP4. The intracellular signaling pathways which include Erk and Akt have been shown to regulate EMT in melanoma, breast cancer, cervical cancer and squamous cell carcinoma [41–44]. Hence it is conceivable that the migration, invasion by IGFBP4 may be due to regulation of EMT phenotype of glioma cells. The role of nuclear localized IGFBP4 is enigmatic. Initially, we hypothesized that the pro-tumorigenic actions of IGFBP4 could be due to nuclear IGFBP4. However, there is differential nuclear localization of IGFBP4 in the two glioma cell lines that were used for over expression or down-regulation (Figure S1). U343 showed cytoplasmic localization of IGFBP4 and U251 showed predominant nuclear localization. It should be noted that IGFBP4 lacks nuclear localization signal [45] and its nuclear localization may depend on other factors. Hence, the factor that facilitates nuclear transport of this protein may be differentially expressed between these cell lines. Interestingly, both these cell lines are pro invasive and that was partly due to IGFBP4 expression. Hence, we conclude that nuclear localization of IGFBP4 may not contribute to the pro-tumorigenic actions of IGFBP4. This opens a new dimension on the role played by IGFBP4 in the nucleus.
In conclusion, our data for the first time advocate a pro-tumorigenic role for IGFBP4 in the progression of GBM pathogenesis. Our previous data and from the literature on the pro-tumorigenic roles by other IGFBP isoforms collectively suggest that IGFBP isoforms 2–5 and 7 may contribute to the pathogenesis of glioma. Hence, a comprehensive approach to interfere with these molecules may provide additional therapeutic options in the management of GBM patients.
References
Pollak MN, Schernhammer ES, Hankinson SE (2004) Insulin-like growth factors and neoplasia. Nat Rev Cancer 4:505–518
LeRoith D, Roberts CT Jr (2003) The insulin-like growth factor system and cancer. Cancer Lett 195:127–137
Maki RG (2010) Small is beautiful: insulin-like growth factors and their role in growth, development, and cancer. J Clin Oncol Off J Am Soc Clin Oncol 28:4985–4995
Foulstone E, Prince S, Zaccheo O, Burns JL, Harper J, Jacobs C, Church D, Hassan AB (2005) Insulin-like growth factor ligands, receptors, and binding proteins in cancer. J Pathol 205:145–153
Fukushima T, Kataoka H (2007) Roles of insulin-like growth factor binding protein-2 (IGFBP-2) in glioblastoma. Anticancer Res 27:3685–3692
Zumkeller W (2001) IGFs and IGFBPs: surrogate markers for diagnosis and surveillance of tumour growth? Mol Pathol 54:285–288
Jones JI, Clemmons DR (1995) Insulin-like growth factors and their binding proteins: biological actions. Endocr Rev 16:3–34
Firth SM, Baxter RC (2002) Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev 23:824–854
Santosh V, Arivazhagan A, Sreekanthreddy P, Srinivasan H, Thota B, Srividya MR, Vrinda M, Sridevi S, Shailaja BC, Samuel C, Prasanna KV, Thennarasu K, Balasubramaniam A, Chandramouli BA, Hegde AS, Somasundaram K, Kondaiah P, Rao MR (2010) Grade-specific expression of insulin-like growth factor-binding proteins-2, -3, and -5 in astrocytomas: IGFBP-3 emerges as a strong predictor of survival in patients with newly diagnosed glioblastoma. Cancer Epidemiol Biomarkers Prev Publ Am Assoc Cancer Res Cospons Am Soc Prev Oncol 19:1399–1408
Jiang W, Xiang C, Cazacu S, Brodie C, Mikkelsen T (2008) Insulin-like growth factor binding protein 7 mediates glioma cell growth and migration. Neoplasia 10:1335–1342
McDonald KL, O’Sullivan MG, Parkinson JF, Shaw JM, Payne CA, Brewer JM, Young L, Reader DJ, Wheeler HT, Cook RJ, Biggs MT, Little NS, Teo C, Stone G, Robinson BG (2007) IQGAP1 and IGFBP2: valuable biomarkers for determining prognosis in glioma patients. J Neuropathol Exp Neurol 66:405–417
Holmes KM, Annala M, Chua CY, Dunlap SM, Liu Y, Hugen N, Moore LM, Cogdell D, Hu L, Nykter M, Hess K, Fuller GN, Zhang W (2012) Insulin-like growth factor-binding protein 2-driven glioma progression is prevented by blocking a clinically significant integrin, integrin-linked kinase, and NF-kappaB network. Proc Natl Acad Sci USA 109:3475–3480
Wetterau LA, Moore MG, Lee KW, Shim ML, Cohen P (1999) Novel aspects of the insulin-like growth factor binding proteins. Mol Genet Metab 68:161–181
Ning Y, Schuller AG, Conover CA, Pintar JE (2008) Insulin-like growth factor (IGF) binding protein-4 is both a positive and negative regulator of IGF activity in vivo. Mol Endocrinol 22:1213–1225
Noll K, Wegmann BR, Havemann K, Jaques G (1996) Insulin-like growth factors stimulate the release of insulin-like growth factor-binding protein-3 (IGFBP-3) and degradation of IGFBP-4 in nonsmall cell lung cancer cell lines. J Clin Endocrinol Metab 81:2653–2662
Babajko S, Leneuve P, Loret C, Binoux M (1997) IGF-binding protein-6 is involved in growth inhibition in SH-SY5Y human neuroblastoma cells: its production is both IGF- and cell density-dependent. J Endocrinol 152:221–227
Wegmann BR, Schoneberger HJ, Kiefer PE, Jaques G, Brandscheid D, Havemann K (1993) Molecular cloning of IGFBP-5 from SCLC cell lines and expression of IGFBP-4, IGFBP-5 and IGFBP-6 in lung cancer cell lines and primary tumours. Eur J Cancer 29A:1578–1584
Pavelic J, Pavelic L, Karadza J, Krizanac S, Unesic J, Spaventi S, Pavelic K (2002) Insulin-like growth factor family and combined antisense approach in therapy of lung carcinoma. Mol Med 8:149–157
Pekonen F, Nyman T, Ilvesmaki V, Partanen S (1992) Insulin-like growth factor binding proteins in human breast cancer tissue. Cancer Res 52:5204–5207
Michell NP, Langman MJ, Eggo MC (1997) Insulin-like growth factors and their binding proteins in human colonocytes: preferential degradation of insulin-like growth factor binding protein 2 in colonic cancers. Br J Cancer 76:60–66
Bachrach LK, Nanto-Salonen K, Tapanainen P, Rosenfeld RG, Gargosky SE (1995) Insulin-like growth factor binding protein production in human follicular thyroid carcinoma cells. Growth Regul 5:109–118
Mita K, Zhang Z, Ando Y, Toyama T, Hamaguchi M, Kobayashi S, Hayashi S, Fujii Y, Iwase H, Yamashita H (2007) Prognostic significance of insulin-like growth factor binding protein (IGFBP)-4 and IGFBP-5 expression in breast cancer. Jpn J Clin Oncol 37:575–582
Damon SE, Maddison L, Ware JL, Plymate SR (1998) Overexpression of an inhibitory insulin-like growth factor binding protein (IGFBP), IGFBP-4, delays onset of prostate tumor formation. Endocrinology 139:3456–3464
Durai R, Yang SY, Seifalian AM, Goldspink G, Winslet MC (2007) Role of insulin-like growth factor binding protein-4 in prevention of colon cancer. World J Surg Oncol 5:128
Ueno K, Hirata H, Majid S, Tabatabai ZL, Hinoda Y, Dahiya R (2011) IGFBP-4 activates the Wnt/beta-catenin signaling pathway and induces M-CAM expression in human renal cell carcinoma. Int J Cancer (Journal international du cancer) 129:2360–2369
van den Boom J, Wolter M, Kuick R, Misek DE, Youkilis AS, Wechsler DS, Sommer C, Reifenberger G, Hanash SM (2003) Characterization of gene expression profiles associated with glioma progression using oligonucleotide-based microarray analysis and real-time reverse transcription-polymerase chain reaction. Am J Pathol 163:1033–1043
Ranganathan P, Agrawal A, Bhushan R, Chavalmane AK, Kalathur RK, Takahashi T, Kondaiah P (2007) Expression profiling of genes regulated by TGF-beta: differential regulation in normal and tumour cells. BMC Genom 8:98
Reddy SP, Britto R, Vinnakota K, Aparna H, Sreepathi HK, Thota B, Kumari A, Shilpa BM, Vrinda M, Umesh S, Samuel C, Shetty M, Tandon A, Pandey P, Hegde S, Hegde AS, Balasubramaniam A, Chandramouli BA, Santosh V, Kondaiah P, Somasundaram K, Rao MR (2008) Novel glioblastoma markers with diagnostic and prognostic value identified through transcriptome analysis. Clin Cancer Res Off J Am Assoc Cancer Res 14:2978–2987
Thangjam GS, Agarwal P, Khan I, Verma UP, Balapure AK, Rao SG, Kondaiah P (2009) Transglutaminase-2 regulation by arecoline in gingival fibroblasts. J Dent Res 88:170–175
Thangjam GS, Kondaiah P (2009) Regulation of oxidative-stress responsive genes by arecoline in human keratinocytes. J Periodontal Res 44:673–682
Pan H, Wang H, Zhu L, Mao L, Qiao L, Su X (2011) The role of Nrf2 in migration and invasion of human glioma cell U251. World Neurosurg 80(3–4):363–370
Zhou R, Flaswinkel H, Schneider MR, Lahm H, Hoeflich A, Wanke R, Wolf E (2004) Insulin-like growth factor-binding protein-4 inhibits growth of the thymus in transgenic mice. J Mol Endocrinol 32:349–364
Singh P, Dai B, Yallampalli U, Lu X, Schroy PC (1996) Proliferation and differentiation of a human colon cancer cell line (CaCo2) is associated with significant changes in the expression and secretion of insulin-like growth factor (IGF) IGF-II and IGF binding protein-4: role of IGF-II. Endocrinology 137:1764–1774
Singh P, Dai B, Dhruva B, Widen SG (1994) Episomal expression of sense and antisense insulin-like growth factor (IGF)-binding protein-4 complementary DNA alters the mitogenic response of a human colon cancer cell line (HT-29) by mechanisms that are independent of and dependent upon IGF-I. Cancer Res 54:6563–6570
Culouscou JM, Shoyab M (1991) Purification of a colon cancer cell growth inhibitor and its identification as an insulin-like growth factor binding protein. Cancer Res 51:2813–2819
Frasca F, Pandini G, Sciacca L, Pezzino V, Squatrito S, Belfiore A, Vigneri R (2008) The role of insulin receptors and IGF-I receptors in cancer and other diseases. Arch Physiol Biochem 114:23–37
Mohan S, Nakao Y, Honda Y, Landale E, Leser U, Dony C, Lang K, Baylink DJ (1995) Studies on the mechanisms by which insulin-like growth factor (IGF) binding protein-4 (IGFBP-4) and IGFBP-5 modulate IGF actions in bone cells. J Biol Chem 270:20424–20431
Cheung PT, Smith EP, Shimasaki S, Ling N, Chernausek SD (1991) Characterization of an insulin-like growth factor binding protein (IGFBP-4) produced by the B104 rat neuronal cell line: chemical and biological properties and differential synthesis by sublines. Endocrinology 129:1006–1015
Dunlap SM, Celestino J, Wang H, Jiang R, Holland EC, Fuller GN, Zhang W (2007) Insulin-like growth factor binding protein 2 promotes glioma development and progression. Proc Natl Acad Sci USA 104:11736–11741
Mendes KN, Wang GK, Fuller GN, Zhang W (2010) JNK mediates insulin-like growth factor binding protein 2/integrin alpha5-dependent glioma cell migration. Int J Oncol 37:143–153
Weiss MB, Abel EV, Mayberry MM, Basile KJ, Berger AC, Aplin AE (2012) TWIST1 is an ERK1/2 effector that promotes invasion and regulates MMP-1 expression in human melanoma cells. Cancer Res 72:6382–6392
Wallin JJ, Guan J, Edgar KA, Zhou W, Francis R, Torres AC, Haverty PM, Eastham-Anderson J, Arena S, Bardelli A, Griffin S, Goodall JE, Grimshaw KM, Hoeflich KP, Torrance C, Belvin M, Friedman LS (2012) Active PI3K pathway causes an invasive phenotype which can be reversed or promoted by blocking the pathway at divergent nodes. PLoS One 7:e36402
Julien S, Puig I, Caretti E, Bonaventure J, Nelles L, van Roy F, Dargemont C, de Herreros AG, Bellacosa A, Larue L (2007) Activation of NF-kappaB by Akt upregulates Snail expression and induces epithelium mesenchyme transition. Oncogene 26:7445–7456
Li J, Zhou BP (2011) Activation of beta-catenin and Akt pathways by Twist are critical for the maintenance of EMT associated cancer stem cell-like characters. BMC Cancer 11:49
Yoshida N, Omoto Y, Inoue A, Eguchi H, Kobayashi Y, Kurosumi M, Saji S, Suemasu K, Okazaki T, Nakachi K, Fujita T, Hayashi S (2004) Prediction of prognosis of estrogen receptor-positive breast cancer with combination of selected estrogen-regulated genes. Cancer Sci 95:496–502
Acknowledgments
We gratefully acknowledge the help of Neurosurgeons Drs. B.S.Chandramouli, A.S. Hedge, A. Arivazhagan, and K. Prasanna for the glioma tissues. We thank Profs. Kumaravel Somasundaram and M.R.S. Rao for useful suggestions during the course of the study. We thank Sarwat Naz for help with ICC, Meenakshi for confocal microscopy facility, Sreekanth Reddy, Harish Srinivasan for RT-PCR data analysis, B.S. Shailaja, Vrinda, Lakshmi and Cini Samuel for help with the patient sample collection. Department of Biotechnology (DBT), India. Infrastructure support was from Department of Science and Technology (under FIST program), DBT, and University Grants Commission (DRS) are acknowledged. VRPK received DBT post-doctoral fellowship.
Conflict of interest
All the Authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11060_2013_1324_MOESM2_ESM.tif
Supplementary material 2 Fig. S1: IGFBP4 expression in U343 and U251 cells upon IGFBP4 over expression and knockdown respectively. a, U343 and b U251 cells were plated on coverslips and allowed to grow. After transfection cells were fixed, permeabilized, stained for IGFBP4 expression. Expression of IGFBP4 is shown in green. Nucleus was stained using propidium iodide (PI), shown in red. Original magnification was 63X. (TIFF 4438 kb)
11060_2013_1324_MOESM3_ESM.tif
Supplementary material 3 Fig. S2: Gene expression changes upon IGFBP4 over expression in U343 cells (a) and knockdown in U251 cells (b). a, Cells were plated and 24 h later transfected with pCMV6-AC-IGFBP4and/or vector. 48 h post transfection, RNA was extracted and gene expression was analyzed by Semi quantitative RT-PCR analysis. b, Cells were plated and 24 h later transfected with IGFBP4 siRNA and/or control siRNA. 96 h post transfection, RNA was extracted and gene expression was analyzed by Semi quantitative RT-PCR analysis. Representative ethidium bromide gel shows the expression of genes regulated upon forced expression of IGFBP4 in U343 (a) and upon IGFBP4 knockdown in U251 glioma cells (b). RPL35a and HPRT expression served as normalizing genes. (TIFF 6968 kb)
Rights and permissions
About this article
Cite this article
Praveen Kumar, V.R., Sehgal, P., Thota, B. et al. Insulin like growth factor binding protein 4 promotes GBM progression and regulates key factors involved in EMT and invasion. J Neurooncol 116, 455–464 (2014). https://doi.org/10.1007/s11060-013-1324-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-013-1324-y