Abstract
Although European beech (Fagus sylvatica L.) is one of the most widespread and ecologically and commercially most important deciduous trees in Europe, little is known about its adaptive genetic variation. We explored single-nucleotide polymorphism (SNP) variation in candidate genes for budburst and drought resistance in beech populations sampled across most of the distribution range, represented in an international provenance trial. SNP variation was monitored for six candidate genes, in 114 individuals from 19 natural populations. Population structure was deduced from the analysis of 20 nuclear microsatellite markers. Different methods to detect imprints of natural selection were used (F ST-outlier, SNP-climate regression, association tests). The F ST-outlier approach identified the COV gene with unambiguous signal of selection, which is an orthologue of Arabidopsis gene for a membrane protein previously reported as phenology-related. Based on environmental association analysis at the population level, the dehydrin gene was found associated with drought-related climatic variables. At the individual level, dehydrin gene also showed a significant association with chlorophyll fluorescence parameters, which are considered stress markers. The importance of the knowledge of physiological variation and geographical patterns of adaptive genetic variation for guiding reproductive materials transfer under climate change is stressed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Local adaptation is a key concept of evolutionary biology (Kawecki and Ebert 2004; Savolainen et al. 2007) with direct implications for forestry. Differences in genotype fitness lead to shifts in genetic structures of populations, which in turn are expected to produce phenotypes providing advantage under the local environment in the offspring generations. There is a plenty of natural and human-induced environmental factors inducing local adaptation in plants, including climate, soil, industrial pollution or biotic agents (mycorrhizas, pollinators, pests), which ultimately lead to changes of the phenotypic expression of a variety of selection-responsive traits (Geburek 2000; Linhart and Grant 1996; Mátyás 1996).
As mentioned above, the issue of local adaptation is of fundamental importance in forestry, especially in relation to the legislation on forest reproductive material (FRM) and the problem of adaptation to the ongoing climate change. One of the main components of the current EU regulations on FRM is the delineation of regions of provenance, serving as a guiding framework for the choice of appropriate reforestation material. Region of provenance is defined as ‘the area or group of areas subject to sufficiently uniform ecological conditions in which stands or seed sources showing similar phenotypic or genetic characters are found’ (European Communities 1999), and may be further subdivided into smaller units according to altitude. In some countries, national regulations prohibit FRM transfer across the boundaries of regions of provenance, especially within the species’ natural ranges. The idea underlying this more or less geography-based approach is that local adaptation is the most important ecological force shaping genetic variation, while climate and photoperiod (i.e., factors associated with the geographical location) are the main drivers of natural selection. Seed sources and reforestation sites within a region of provenance are expected to share similar environments. In contrast, non-local seed sources are expected to be genetically divergent; transfer of FRM from distant regions of provenance may be risky because of possible losses in terms of survival, yield and other relevant benefits (Hemery 2008). However, there is no unanimity about the reasonable size or number of regions of provenance among countries, neither there is a consensus about the principles and methodologies they should be based on. Empirical data sometimes do not provide any support for the existing zones (de la Mata and Zas 2010).
At present, these transfer rules are questioned in association with climate change. Assisted migration is proposed as a solution (Williams and Dumroese 2013), i.e., transfer of genetic material from populations, which in the past have experienced climatic conditions expected on target sites in the future. The logic of this measure also relies on climate-driven local adaptation.
Most information on adaptation and patterns of adaptive genetic variation has come from field experiments until now. A traditional tool for the study of adaptation in forest trees are provenance trials (or combined provenance–progeny tests), which represent a specific type of common-garden experiments, widely used in the studies of plant evolution and ecology (Briggs and Walters 1997). The term “provenance” denotes a local subpopulation of a defined origin. Modern provenance experiments have frequently been established on a large scale, i.e., numerous provenances frequently covering a substantial part of the range of a species were repeatedly planted on series of trial plots under a wide range of environments. Such experimental setups allow testing the reactions of provenances to transfer; on the one hand, determining the environment where a particular provenance performs best, on the other hand, choosing the optimum provenance for a particular site (not necessarily the local one). There are several methodological approaches that allow making inference on the extent and patterns of local adaptation: general transfer functions and population response functions (Rehfeldt et al. 1999) and comparisons of differentiation at fitness-related traits vs. neutral markers (Q ST vs. F ST; Merilä and Crnokrak 2001) are the most commonly used ones.
The information on the adaptation processes and geographic patterns of genetic variation produced by adaptation at the molecular level is by far not as rich, in spite of recent rapid developments in forest tree genomics (González-Martínez et al. 2006; Grattapaglia et al. 2009; Neale and Ingvarsson 2008; Plomion et al. 2016). Candidate gene approach has been preferred in genomic studies of forest trees, as the rapid decay of linkage disequilibrium in tree populations makes association studies difficult (Neale and Kremer 2011). Even though reference genomes for tree species gradually appear, the choice of candidate genes is mostly based on orthology or functionally equivalency to genes identified in model species (mainly Arabidopsis, rice, poplar), and in most cases genes related to climatic adaptation such as those underlying phenology and drought or heat resistance are targeted. Once validated by gene expression studies, QTL detection or association tests, these genes are used in the studies of natural populations to assess patterns of adaptive genetic variation (Neale and Kremer 2011). Polymorphisms associated with phenology, drought resistance and other climate-related traits were identified in European beech or other Fagaceae; however, they were mostly tested in local-scale experiments (Derory et al. 2006; Lalagüe et al. 2014; Müller et al. 2015; Rellstab et al. 2016; Seifert et al. 2012). There is still a general lack of empirical studies focusing on the relationship between genetic structures of populations at (putatively) adaptive loci with their environments done on larger scales (but see Eckert et al. 2010; Mosca et al. 2014; Scalfi et al. 2014; Wachowiak et al. 2009). Identification of adaptive variation trends and patterns is moreover complicated by the fact that genetic structures at loci under selection are influenced by neutral processes such as gene flow, migration or genetic drift; disentangling the role of these two types of processes is not a simple task (Chen et al. 2012; Grivet et al. 2011).
Because of their long generation time, long-lived forest trees are exposed to environmental (especially climatic) fluctuations during ontogeny. Phenotypic plasticity, which allows avoiding selection pressures at least in some extent (Mimura and Aitken 2010), may thus become the preferred strategy for coping with environmental heterogeneity (Vitasse et al. 2009). Moreover, pollen dispersal in trees is generally very efficient, especially in wind-pollinated species, and may prevent differentiation (Savolainen et al. 2007). European beech is a typical representative of wind-pollinated forest tree species with a large and relatively continuous distribution range (cf. http://www.euforgen.org/distribution-maps/) comprising climatically and edaphically heterogeneous environments. In contrast to other temperate trees in Europe, beech colonized major part of its present range from a single refugium, located close to the eastern foothills of the Alps (Slovenia, Istria; Magri et al. 2006). This does not eliminate all possible influences of historical and demographical factors on genetic variation patterns, but populations sharing refugial origin also share a substantial part of their initial genetic pools, which were then subjected to selection pressures imposed by climate, soil, competitors and other sources of environmental stress. In this sense, such situation simplifies adaptation studies considerably, because a part of false signals of selection resulting from demographic processes is eliminated. Our study focuses on signs of local adaptation within a set of provenances represented in the Slovak trial of an international provenance experiment. Our objectives were (1) testing which particular single-nucleotide polymorphisms in candidate genes for phenology and drought stress identified in earlier studies as phenology- and drought stress-related are affected by selection, (2) identification of their relationships with climatic variables and (3) identification of their potential associations with vegetative phenology and physiological traits.
Materials and methods
Experimental material
The study relies on an international provenance experiment with European beech established under the auspices of the Institute of Forest Genetics (Johann Heinrich von Thünen Institute) in Grosshansdorf, Germany in 1998 (seed sowing 1996). The Slovak trial plot was planted at the locality Tále near Zvolen, central Slovakia (48°38′N, 19°02′E, 810 m a.s.l.) in a former forest nursery and comprises 32 provenances covering a major part of the distribution range of beech in Europe. The test was established under a randomized complete block design with three blocks, each plot containing initially 50 plants planted at 2 × 1-m spacing. For this study, a subset of 19 provenances was used (see Suppl. 1), with a preference of populations originated from the Slovenian glacial refugium to minimize the effects of history and demography on genetic variation (cf. Magri et al. 2006). As an outgroup population, the provenance Lagast (Massif Central, France) was used, which probably has originated from a refugium located in southeast France (Massif Central, Rhône valley).
Climatic variables of the sites the provenances originated were taken from the database maintained by the provenance experiment coordinator (provided by the material suppliers) and complemented by bioclimatic variables from the WorldClim high-resolution interpolated climate database: BIO4 (temperature seasonality), BIO5 (maximum temperature of the warmest month), BIO6 (minimum temperature of the coldest month), BIO7 (temperature annual range), BIO13 (precipitation of the wettest month), BIO14 (precipitation of the driest month) and BIO15 (precipitation seasonality). The bioclimatic variables are derived from meteorological data within the period 1960–1990 at the 30″ resolution.
Growth and phenological traits
Several traits were assessed within independent series of measurements during the period 2007–2013. The assessment of growth traits (height, diameter at breast height), phenology and late-frost damage was done exhaustively. Height and diameter growth were measured in 2007 and 2013, vegetative phenology in 2007, 2008 and 2012, and late-frost damage after a severe frost event on May 1, 2007. Details of the assessment of budburst and leaf-discoloration phenology (scoring scales, scoring dates, estimation procedure for the Julian middays of spring flushing and autumn discoloration, respectively) and frost-damage scoring are given in Gömöry and Paule (2011).
Physiological traits
As photosynthesis reacts sensitively to overheating and water stress, the choice of the scored physiological parameters focused on various aspects of photosynthetic processes.
Assessments of chlorophyll a fluorescence and gas exchange parameters were done during the summer seasons 2013 and 2014. The following parameters were assessed:
Fast kinetics of chlorophyll a fluorescence: the maximum fluorescence yield (F v/F m), photosynthetic performance index (PI), density of active photosystem II reaction centers (RC/ABS) and relative variable fluorescence at the K-step of the fluorescence induction curve (Wk). Thermal stability of photosystem II: T 15, the temperature at which F v/F m declines 15% from the maximum value.
Slow kinetics of chlorophyll a fluorescence: the actual photochemical efficiency of the open reaction centers of PSII (Φ PSII); the maximum Φ PSII (Φ PSII(max)) and the area below the response curve (Φ PSII(Area)) were assessed.
Gas exchange CO2 assimilation, stomatal conductance, transpiration rate and the instantaneous water-use efficiency (WUE).
Details of the assessment procedures are described in Gömöry et al. (2015) and Suppl. 2a.
DNA extraction, microsatellite analysis and sequencing
As a rule, leaves were collected from six trees per population (one leaf from the upper part of the crown per tree). For microsatellite analyses, larger sample sizes were used for four populations (115, 120, 115 and 108 trees of the provenances 15, 48, 51 and 67, respectively; see Suppl. 1) to get a more reliable information about neutral variation. Total genomic DNA was extracted from the silica-dried leaves using a modified CTAB protocol following Doyle and Doyle (1987).
Polymorphisms in six candidate genes for phenology and drought stress (expected to be responsive to climatic contrasts) were analyzed (Table 1). Primer sequences and thermal cycling profile for PCR followed Seifert et al. (2012) and Lalagüe et al. (2014). The PCR mixtures for all markers were done in volume 20 μl consisting of 1× PCR buffer, 2 mM MgCl2, 0.2 µM of primer, 0.3 µM dNTP, 0.5 U Taq DNA polymerase (GeneCraft), 0.8 μg/μl of BSA and 25 ng of template DNA. The PCR products were checked on 1.5% agarose gel and afterward they were sent to IGA Technology Services (Udine, Italy) for sequencing. For all primer pairs, both DNA strands were sequenced. Obtained raw data were evaluated using SeqScape v.2.5.
In addition, twenty microsatellite markers according to Lefèvre et al. (2012), Tanaka et al. (1999) and Pastorelli et al. (2003) were studied. For details on PCR and scoring, see Gömöry et al. (2015) and Suppl. 2b.
Data analysis
Sequences were reduced to sites exhibiting single-nucleotide polymorphisms (SNPs). For statistical evaluations, each polymorphic site was treated as separate locus. Phase (i.e., combination of alleles present on each of the two chromosomes) was reconstructed using the Bayesian MCMC algorithm of Stephens and Donelly (2003). The program DnaSP v5 (Librado and Rozas 2009) was used.
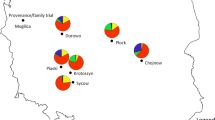
To reveal potential substructure both in the nSSR and sequence data, we applied the Bayesian clustering algorithm implemented in STRUCTURE version 2.3.4 (Pritchard et al. 2000a) to cluster individuals into K groups, employing the admixture model with correlated allele frequencies. K values were tested from 1 to 8. A burn-in period of 500,000 iterations was followed by 1,000,000 iterations for estimation of the membership coefficients. Ten independent Markov chains were run for each K. The choice of the true number of clusters K was based on the method of Evanno et al. (2005), implemented in STRUCTURE HARVESTER (Earl and von Holdt 2012), and the run with the highest posterior probability for the selected K was chosen (Suppl. 3a, 3b).
Isolation by distance (IBD) was tested using the regression method of Rousset (1997). Because IBD patterns can be distorted by selection, the analysis was done separately for putatively neutral nSSR markers and potentially selected SNPs. Geographical coordinates were converted to orthogonal coordinates using Lambert conformal conic projection (http://twcc.free.fr/). As we have no exact information about paths of migration, simple Euclidean distances between populations were calculated. As recommended for a two-dimensional stepping-stone model, estimates of F ST/(1 − F ST) were regressed against logarithm of distance, whereas the significance of the slope was tested by a Mantel test with 99,999 permutations (program Genepop; Rousset 2008). As stated by Rousset (1997), differentiation among samples at short distances deviates from the expectations of the two-dimensional stepping-stone model; therefore, population pairs separated by less than 50 km were omitted from the analysis. Exact tests of allelic differentiation among populations were also performed in Genepop using 10,000 dememorization steps followed by 1000 batches of 5000 iterations each, and the resulting probabilities were corrected using sequential Bonferroni correction.
Frequency-based detection of outlier loci was performed using two approaches. First, we used Bayescan (Foll and Gaggiotti 2008); the fact that all studied populations have originated from the same Slovenian refugium (the population Lagast was excluded from this analysis) complies with the null model of Bayescan, which assumes a common migrant genepool. Because all SNPs were located in candidate genes and can thus be at least potentially under selection, we included the scored nSSR markers as a kind of “neutral standard.” Bayescan v. 2.1 was used for the computations, with 20 pilot runs of 5000 iterations each and a burn-in of 50,000 iterations followed by 50,000 iterations for the estimation of the posterior distributions with a thinning interval of 10. Prior odds for the neutral model were set to 10 (default). The evidence of selection was based on Bayes factors, measuring odds for selection model versus neutral model derived from posterior probabilities of each of the models. Posterior odds of more than 32:1 in favor of the selection model (i.e., log10(odds) >1.5) is taken as a strong evidence for selection (Foll and Gaggiotti 2008). False discovery rate (FDR; the expected proportion of false positives among outlier markers) was controlled through the q value, quantifying the minimum FDR at which this locus may become significant (q value is defined in the context of multiple testing across all polymorphic loci). Second, we used the F ST-outlier method of Beaumont and Nichols (1996) (Fdist) as applied in the software Lositan (Antao et al. 2008), where the neutral envelope for F ST across loci was obtained from 500,000 simulations under the finite island model. As microsatellites are expected to follow stepwise mutation model (in contrast to SNPs), the simulations were done with the SNP dataset only. F ST values conditioned on heterozygosity and outside the 99% confidence interval were considered candidate outliers.
Climatic variables were tested for correlations with SNP allele frequencies employing multiple univariate logistic regression models with the spatial analysis method (SAM), as implemented in the Samβada v0.5.1 software (Joost et al. 2007). The significance of correlations was assessed using the likelihood-ratio test and the Wald test implemented in SAM, applying an initial 95% confidence interval. As the SAM requires input of the presence/absence data, genotypes were coded as suggested by Joost et al. (2007) (the effect of a particular SNP allele controlling putative environment-related traits is expected to be dominant). Sequential Bonferroni correction for multiple testing was applied; correlations that remained significant after correction for both test statistics were retained. In addition, Bayesian factors for the support for models in which SNPs covary linearly with environmental variables over models in which SNPs vary according to neutral expectation were assessed using BayEnv2 (Coop et al. 2010). For each SNP, BayEnv2 was run with 100,000 iterations and 15 environmental variables.
Associations between SNPs and the phenotypic traits were assessed using TASSEL v. 5.0 (Bradbury et al. 2007). As a previous study based on the same material (Gömöry et al. 2015) revealed absence of any kinship structure (all trees within provenances are unrelated), we tested the SNP/trait associations under the general linear model (GLM):
where y is the vector of phenotypic data), X is the design matrix, b is a vector of fixed effects (SNP and the population structure) and e is the vector of random residues. The population structure (Q matrix) was deduced from the STRUCTURE analysis of neutral nSSR markers (see above).
Results
In total, 46 SNPs were identified. Sites with overall minor allele frequency below 2% were discarded from further evaluations.
Isolation by distance proved to be significant although not strong, when estimated both by nSSR and SNP loci (Spearman’s rank correlation coefficient between F ST/(1 − F ST) and log(distance) was 0.330 P = 0.0004 and 0.297 P = 0.0031 for nSSR and SNP, respectively). Regression slopes are slightly different (0.015 (95% CI 0.005–0.027) and 0.025 (95% CI 0.016–0.035) for nSSR and SNP, respectively), but their permutation-based confidence intervals overlap considerably.
In the analysis of the SNP dataset under the STRUCTURE procedure, the ΔK measure (Evanno et al. 2005) indicated K = 2 as the most probable number of groups (Suppl. 3a). However, it must be reminded that by principle, ΔK does not allow inference on K = 1, i.e., absence of any structure. The distribution of cluster proportions does not follow any geographical trend and shows no association with any phenotypic trait or climatic variable except maximum July temperature (BIO5). On the other hand, neutral markers showed clear geographic trends at K = 3 and, consequently, significant correlations with geographically patterned climatic variables and phenotypic traits such as phenology (Suppl. 4).
None of the nSSR loci showed significant differentiation. On the other hand, among 46 polymorphic sites in the candidate genes, five exhibited significant differences in allele frequencies even after the sequential Bonferroni correction (Table 2, Suppl. 5).
Both F ST-outlier-based approaches to identification of polymorphisms subjected to selection yielded consistent results: signs of divergent selection were detected for two closely linked SNPs in the locus F (Table 2). The log (posterior odds) value was 1.8 and 2.3, respectively, which means a very strong to decisive support for selection (Foll and Gaggiotti 2008). The q values (minimum FDR) were very low, suggesting that these SNPs are not false positives.
The two methods relating the SNPs with climates at the sites of origin did not yield consistent outcomes. BayEnv failed to identify any signals for SNP/climate relationship (Suppl. 6). On the other hand, SAM revealed the C/G polymorphism at the site 842 of the dehydrin gene (marker B) as correlated with drought-related variables: the Ellenberg quotient, annual precipitation total and precipitation of the driest month (BIO14). Moreover, there is a certain (although weaker) signal of temperature- and precipitation-driven selection at sites 454 and 475 of the histone h3-gene (marker E; C/T and A/C, respectively) (Table 3, Suppl. 7).
Using the Q matrix resulting from the STRUCTURE analysis (K = 3) as covariates, a total of six SNP–trait associations were identified, which remained significant after sequential Bonferroni correction (Table 4, Suppl. 8). Phenotypic variation explained by the marker–trait associations was quite low, ranging from 3.8 to 7.1%. Most significant associations were observed for chlorophyll fluorescence traits reflecting the state of the photosynthetic apparatus, and in one case for a growth trait (height); surprisingly, no association was observed for phenological traits.
Discussion
Population structure
Although beech is an anemophilous species having small pollen grains with high motility, it is known to develop isolation-by-distance patterns both on the local (Chybicki et al. 2009) and regional (Gömöry et al. 2007) scales. This was confirmed by our study, which may pose a problem for the interpretation of outlier-based identification of the signals of selection, as isolation by distance may yield false positive results (Lotterhos and Whitlock 2014). In our case, IBD was significant but weak in case of both neutral and potentially adaptive markers; the regression slopes as well as correlation coefficients between genetic and geographic distances were low. In spite of a nonsignificant difference between the slopes, the fact that the slope is bigger in the case of SNPs (i.e., differentiation increases more rapidly with distance compared to nSSRs) may indicate that non-neutral processes contributed to differentiation.
Another source of false positives in selection tests may be recent bottlenecks (Foll and Gaggiotti 2008), which we did not test explicitly because of small sample sizes in many provenances. However, beech is a species with generally continuous range. The provenance trial under the study focused on typical representatives of beech populations and does not include isolated fragments with limited population size which may be suspect of genetic bottleneck. This, of course, does not rule out traces of founder effect in peripheral populations at the front edge of the Holocene recolonization (Comps et al. 2001). Our study includes populations from the northern distribution margin but, judging from the STRUCTURE analysis, they are not excessively differentiated compared to the rest. This is not a proof of the absence of a bottleneck but decreases the probability that any of the studied provenances experienced a recent reduction in population size.
Bayesian analysis of population structure based on nuclear microsatellites, which are generally considered neutral, revealed close correlations between genetic cluster proportions in provenance gene pools and geographical coordinates and/or some climatic variables. Very probably this reflects a specific colonization pattern of beech. In contrast to most temperate European tree species, European beech colonized most of its present distribution area from a single refugium, positioned quite eccentrically in the Southeast (Magri et al. 2006). The studied provenance experiment (series 1996/1998) comprises very few populations originating from the other refugia. Consequently, population structure of the provenances involved in this study is a product of northward and westward migration reflected in significant correlations of cluster proportions with latitude and longitude (cf. Suppl. 4). A significant correlation with altitude is a sampling artifact: altitude is collinear with geographical coordinates, high-elevation provenances are concentrated in Central Europe near the Slovenian/Istrian refugium, whereas northern and western marginal populations are located close to the sea level. The same is true for climatic variables associated with the geographical position, and also for phenology, which shows a clear longitudinal trend in Europe (von Wuehlisch et al. 1995; Gömöry and Paule 2011). Nevertheless, the existence of geographical and climate-related trends of neutral variation underlines the necessity for considering the population structure in the search for adaptive polymorphisms and phenotype–genotype associations (Bradbury et al. 2007; Pritchard et al. 2000b).
Signals of selection and environmental associations
All markers used in this study were selected based on associations with climate or climate-related phenotypic traits such as drought tolerance or budburst phenology proven or indicated in earlier studies with beech or other Fagaceae (Lalagüe et al. 2014; Vornam et al. 2011). Therefore, we expected positive hits in most markers, taking into account broad climatic gradients and variations in phenotypic traits in the materials that we used.
This expectation was fulfilled only partially. The F ST-outlier approach identified only one candidate gene, namely the COV, with unambiguous signal of selection. The orthologue in Arabidopsis is the gene controlling the synthesis of continuous vascular ring protein, which is a membrane protein localized in trans-Golgi network participating in secretion, vacuolar trafficking and endocytosis (Shirakawa et al. 2014). The gene was reported as phenology-related by Lalagüe et al. (2014) because an earlier study in Quercus petraea revealed a strong association of this locus with budburst in segregating progenies (Derory et al. 2006). The exact way how it is involved in the cascade of genes responsible for spring phenology is, however, unknown. As the TASSEL analysis did not show a significant association of any SNP within this locus with budburst or leaf discoloration, it remains questionable whether phenology-driving environmental factors are indeed the drivers of selection acting at this locus.
Environmental association analysis at the population level rendered the dehydrin gene as associated with aridity gradients. This locus is responsive to abscisic acid and has been shown to be involved in drought response of beech seeds (Jiménez et al. 2008). In general, dehydrins are supposed to protect the cell metabolism during cellular dehydration caused by multiple factors including drought, low temperature, salinity and their expression may also be induced by short daylength (Close 1996; Kosová et al. 2007; Welling et al. 2004). Allelic variation in dehydrin genes may thus reflect selection induced by several different environmental stresses. Nevertheless, all climatic variables showing association with the SNP at this locus are more or less closely related to drought (Ellenberg quotient, July precipitation, annual precipitation), and the observed variation is thus plausibly suggested to reflect adaptation to water stress.
At the individual level, several significant associations were found. Surprisingly, in spite of huge contrasts in spring and autumn phenology among the studied beech provenances (cf. Gömöry and Paule 2011) none of the genes previously reported as related to phenology showed affinity to timing of budburst or leaf discoloration. On the other hand, the dehydrin gene again showed moderate association with chlorophyll fluorescence parameters, which reflect the state of photosystem II reaction centers and, consequently, efficiency of electron transport in PSII. Photoinhibition as a symptom of the damage to PSII is the first manifestation of stress in a leaf (Maxwell and Johnson 2000). As all chlorophyll fluorescence measurements were done after an at least two-weeks-long period of drought during full summer, the most probable sources of stress for beech plants were heat and drought. In this sense, the phenotype–SNP associations are consistent with population-level environmental associations, showing significant outcomes at the same gene.
Another gene showing association with a physiological stress marker was the gene for isocitric dehydrogenase. The distribution of isoforms of this enzyme in the conifer Abies alba is known to be associated with their thermostability (Bergmann and Gregorius 1993), and its activity was shown to be associated with drought stress in Arabidopsis (Gálvez et al. 2005). To what extent this may apply for beech is questionable, explaining the observed association by water or temperature stress based on such analogies would be a pure speculation.
Implications for forestry
Climate change is expected to have significant impact on health, yield potential and sometimes the sole existence of forest tree populations in Europe (Hanewinkel et al. 2013). Assisted migration proposed as the main mitigation measure for forest trees (Williams and Dumroese 2013) requires knowledge of the responses of forest tree populations to such transfer in terms of survival and growth. This information is currently derived from provenance research at the first place. Even though this source must not be underestimated, it has several limitations. In many cases, the autochthony of provenances is questionable; many forests in Europe were established artificially, reproductive materials were frequently transferred over large distances, and tracking of these movements is sometimes hardly feasible because of the lack of records. Provenance trials are commonly measured at ages which are mostly far from maturity, and the extrapolation of results to the older age need not be reliable. Trials conducted under controlled conditions insufficiently reflect biotic interactions; changed occurrence patters of pests and diseases seem to be more important for the health and survival of trees than climatic adaptation as such. And finally, spatial resolution of provenance experiments is too coarse for a reliable selection of seed sources for an uncertain future. A deeper insight into adaptation processes to guide assisted migration is needed than can be provided by field trials. Key stressors have already been identified but physiological processes underlying responses to environment are not completely known. For instance, currently there exist several phenology models which predict dates of budburst with equal or similar accuracy, although they are based on completely different assumptions; a mechanistic model of the timing of budburst as an important adaptive trait is not available (Saxe et al. 2001). In other cases, the problem is more of technical nature—for each relevant physiological process a descriptor needs to be found, a parameter which preferably can be measured on large numbers of individuals in a short time to allow large-scale phenotyping. As for genetic variation, extensive information is available about spatial structuration of the neutral genetic variation, which allowed localization of glacial refugia and provided knowledge on gene flow, founder effects associated with migration and other neutral processes. As neutral processes also affect adaptive variation in tree populations (e.g., Bošeľa et al. 2016), such information is relevant both for the studies of adaptation and the forestry practice in terms of FRM production and use, but is not explicitly linked to selection processes underlying adaptation. On the other hand, the knowledge of adaptive genetic polymorphisms sensu stricto is still rudimentary. In spite of a recent progress, the current state-of-the-art in the case of non-model organisms such as forest trees is generally the same as shown in our study: identification of signals of selection and associations with climatic variables or climate-related traits in the genome, whereas a complex understanding of the genome–climate relationships as the ultimate goal is still far from being reached. Nevertheless, it is just the identification of adaptively relevant polymorphisms and mapping their distribution within species’ ranges, which are necessary for the prediction of responses of tree populations to changing climate and setting reliable guidelines for FRM transfer. The intention of this study was making a step in this direction.
References
Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G (2008) LOSITAN: a workbench to detect molecular adaptation based on a F ST-outlier method. BMC Bioinformatics 9:323. doi:10.1186/1471-2105-9-323
Beaumont MA, Nichols RA (1996) Evaluating loci for use in the genetic analysis of population structure. Proc R Soc Lond B 263:1619–1626
Bergmann F, Gregorius H-R (1993) Ecogeographical distribution and thermostability of isocitrate dehydrogenase (IDH) alloenzymes in European silver fir (Abies alba). Biochem Syst Ecol 21:597–605
Bošeľa M, Popa I, Gömöry D, Longauer R, Tobin B, Kyncl J, Kyncl T, Nechita C, Petráš R, Sidor C, Šebeň V, Büntgen U (2016) Effects of post-glacial phylogeny and genetic diversity on the growth variability and climate sensitivity of European silver fir. J Ecol 104:716–724
Bradbury PJ, Zhang ZW, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Briggs D, Walters SM (1997) Plant variation and evolution, 3rd edn. Cambridge University Press, Cambridge, 521 + xxi pp
Chen J, Källman T, Ma X, Gyllenstrand N, Zaina G, Morgante M, Bousquet J, Eckert A, Wegrzyn J, Neale D, Lagercrantz U, Lascoux M (2012) Disentangling the roles of history and local selection in shaping clinal variation of allele frequencies and gene expression in Norway spruce (Picea abies). Genetics 191:865–881
Chybicki IJ, Trojankiewicz M, Oleksa A, Dzialuk A, Burczyk J (2009) Isolation-by-distance within naturally established populations of European beech (Fagus sylvatica). Botany 87:791–798
Close TJ (1996) Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol Plant 97:795–803
Comps B, Gömöry D, Letouzey J, Thiébaut B, Petit RJ (2001) Diverging trends between heterozygosity and allelic richness during postglacial colonization in the European beech. Genetics 157:389–397
Coop G, Witonsky D, Di Rienzo A, Pritchard JK (2010) Using environmental correlations to identify loci underlying local adaptation. Genetics 185:1411–1423
de la Mata R, Zas R (2010) Transferring Atlantic maritime pine improved material to a region with marked Mediterranean influence in inland NW Spain: a likelihood-based approach on spatially adjusted field data. Eur J For Res 129:645–658
Derory J, Leger P, Garcia V, Schaeffer J, Hauser MT, Salin F, Luschnig C, Plomion C, Glossl J, Kremer A (2006) Transcriptome analysis of bud burst in sessile oak (Quercus petraea). New Phytol 170:723–738
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Earl DA, von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Res 4:359–361
Eckert AJ, Bower AD, González-Martínez SC, Wegrzyn JL, Coop G, Neale DB (2010) Back to nature: ecological genomics of loblolly pine (Pinus taeda, Pinaceae). Mol Ecol 19:3789–3805
European Communities (1999) Council directive 1999/105/EC of 22 December 1999 on the marketing of forest reproductive material. Off J Eur Commun 15. 1. 2000 17–L 11/40
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Foll M, Gaggiotti O (2008) A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180:977–993
Gálvez L, González EM, Arrese-Igor C (2005) Evidence for carbon flux shortage and strong carbon/nitrogen interactions in pea nodules at early stages of water stress. J Exp Bot 56:2551–2561
Geburek T (2000) Effects of environmental pollution on the genetics of forest trees. In: Young A, Boshier D, Boyle T (eds) Forest conservation genetics, principles and practice. CSIRO Publishing, Colingwood and CABI Publishing, Oxon, pp 135–158
Gömöry D, Paule L (2011) Trade-off between height growth and spring flushing in common beech (Fagus sylvatica L.). Ann For Sci 68:975–984
Gömöry D, Paule L, Vyšný J (2007) Patterns of allozyme variation in western-Eurasian beeches. Bot J Linn Soc 154:165–174
Gömöry D, Ditmarová L, Hrivnák M, Jamnická G, Kmeť J, Krajmerová D, Kurjak D (2015) Differentiation in phenological and physiological traits in European beech (Fagus sylvatica L.). Eur J For Res 134:1075–1085
González-Martínez SC, Krutovsky KV, Neale DB (2006) Forest-tree population genomics and adaptive evolution. New Phytol 170:227–238
Grattapaglia D, Plomion C, Kirst M, Sederoff RR (2009) Genomics of growth traits in forest trees. Curr Opin Plant Biol 12:148–156
Grivet D, Sebastiani F, Alia R, Bataillon T, Torre S, Zabal-Aguirre M, Vendramin GG, González-Martínez SC (2011) Molecular footprints of local adaptation in two Mediterranean conifers. Mol Biol Evol 28:101–116
Hanewinkel M, Cullmann DA, Schelhaas M-J, Nabuurs G-J, Zimmermann NE (2013) Climate change may cause severe loss in the economic value of European forest land. Nat Clim Chang 3:203–207
Hemery GE (2008) Forest management and silvicultural responses to projected climate change impacts on European broadleaved trees and forests. Int For Rev 10:591–607
Jiménez JA, Alonso-Ramírez A, Nicolás C (2008) Two cDNA clones (FsDhn1 and FsClo1) up-regulated by ABA are involved indrought responses in Fagus sylvatica L. seeds. J Plant Physiol 165:1798–1807
Joost S, Bonin A, Bruford MW, Després L, Conord C, Erhardt G, Taberlet P (2007) A spatial analysis method (SAM) to detect candidate loci for selection: towards a landscape genomics approach to adaptation. Mol Ecol 16:3955–3969
Kawecki TJ, Ebert D (2004) Conceptual issues in local adaptation. Ecol Lett 7:225–1241
Kosová K, Vítámvás P, Prášil IT (2007) The role of dehydrins in plant response to cold. Biol Plant 51:601–617
Lalagüe H, Csilléry K, Oddou-Muratorio S, Safrana J, de Quattro C, Fady B, González-Martínez SC, Vendramin GG (2014) Nucleotide diversity and linkage disequilibrium at 58 stress response and phenology candidate genes in a European beech (Fagus sylvatica L.) population from southeastern France. Tree Genet Genomes 10:15–26
Lefèvre S, Wagner S, Petit RJ, de Lafontaine G (2012) Multiplexed microsatellite markers for genetic studies of beech. Mol Ecol Res 12:484–491
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Linhart YB, Grant MC (1996) Evolutionary significance of local genetic differentiation in plants. Annu Rev Ecol Syst 27:237–277
Lotterhos KE, Whitlock MC (2014) Evaluation of demographic history and neutral parameterization on the performance of F ST outlier tests. Mol Ecol 23:2178–2192
Magri D, Vendramin GG, Comps B, Dupanloup I, Geburek T, Gömöry D, Latalowa M, Litt T, Paule L, Roure JM, Tantau I, van der Knaap WO, Petit RJ, de Beaulieu J-L (2006) A new scenario for the Quaternary history of European beech populations: palaeobotanical evidence and genetic consequences. New Phytol 171:199–222
Mátyás C (1996) Climatic adaptation of trees: rediscovering provenance tests. Euphytica 92:45–54
Maxwell K, Johnson GN (2000) Chlorophyll fluorescence—a practical guide. J Exp Bot 51:659–668
Merilä J, Crnokrak P (2001) Comparison of genetic differentiation at marker loci and quantitative traits. J Evol Biol 14:892–903
Mimura M, Aitken SN (2010) Local adaptation at the range peripheries of Sitka spruce. J Evol Biol 23:249–258
Mosca E, González-Martínez SC, Neale DB (2014) Environmental versus geographical determinants of genetic structure in two subalpine conifers. New Phytol 201:180–192
Müller M, Seifert S, Finkeldey R (2015) A candidate gene-based association study reveals SNPs significantly associated with bud burst in European beech (Fagus sylvatica L.). Tree Genet Genomes 11:116
Neale DB, Ingvarsson PK (2008) Population, quantitative and comparative genomics of adaptation in forest trees. Curr Opin Plant Biol 11:149–155
Neale DB, Kremer A (2011) Forest tree genomics: growing resources and applications. Nat Rev Genet 12:111–122
Pastorelli R, Smulders MJM, Van’t Westende WPC, Vosman B, Giannini R, Vettori C, Vendramin GG (2003) Characterization of microsatellite markers in Fagus sylvatica L. and Fagus orientalis Lipsky. Mol Ecol Notes 3:76–78
Plomion C, Bastien C, Bogeat-Triboulot MB, Bouffier L, Déjardin A, Duplessis S, Fady B, Heuertz M, Le Gac AL, Le Provost G, Legue V, Lelu-Walter MA, Leplé JC, Maury S, Morel A, Oddou-Muratorio S, Pilate G, Sanchez L, Scotti I, Scotti-Saintagne C, Segura V, Trontin JF, Vacher C (2016) Forest tree genomics: 10 achievements from the past 10 years and future prospects. Ann For Sci 73:77–103
Pritchard JK, Stephens M, Donnelly P (2000a) Inference of population structure from multilocus genotype data. Genetics 155:945–959
Pritchard JK, Stephens M, Rosenberg NA, Donnelly P (2000b) Association mapping in structured populations. Am J Hum Genet 67:170–181
Rehfeldt GE, Ying CC, Spittlehouse DL, Hamilton DA (1999) Genetic responses to climate in Pinus concorta: niche breadth, climate change, and reforestation. Ecol Monogr 69:375–407
Rellstab C, Zoller S, Walthert L, Lesur I, Pluess AR, Graf R, Bodénès C, Sperisen C, Kremer A, Gugerli F (2016) Signatures of local adaptation in candidate genes of oaks (Quercus spp.) with respect to present and future climatic conditions. Mol Ecol 25:5907–5924
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Rousset F (2008) Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Res 8:103–106
Savolainen O, Pyhajärvi T, Knurr T (2007) Gene flow and local adaptation in trees. Ann Rev Ecol Evol Syst 38:595–619
Saxe H, Cannell MGR, Johnsen B, Ryan MG, Vourlitis G (2001) Tree and forest functioning in response to global warming. New Phytol 149:369–399
Scalfi M, Mosca E, Di Pierro EA, Troggio M, Vendramin GG, Sperisen C, La Porta N, Neale DB (2014) Micro- and macro-geographic scale effect on the molecular imprint of selection and adaptation in Norway spruce. PLoS ONE 9:e115499
Seifert S, Vornam B, Finkeldey R (2012) DNA sequence variation and development of SNP markers in beech (Fagus sylvatica L.). Eur J For Res 131:1761–1770
Shirakawa M, Ueda H, Koumoto Y, Fuji K, Nishiyama C, Kohchi T, Hara-Nishimura I, Shimada T (2014) Continuous vascular ring (COV1) is a trans-Golgi network-localized membrane protein required for Golgi morphology and vacuolar protein sorting. Plant Cell Physiol 55:764–772
Stephens M, Donelly P (2003) A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73:1162–1169
Tanaka K, Tsumura Y, Nakamura T (1999) Development and polymorphism of microsatellite markers for Fagus crenata and the closely related species, F. japonica. Theor Appl Genet 99:11–15
Vitasse Y, Delzon S, Bresson CC, Michalet R, Kremer A (2009) Altitudinal differentiation in growth and phenology among populations of temperate-zone tree species growing in a common garden. Can J For Res 39:1259–1269
von Wuehlisch G, Krusche D, Muhs HJ (1995) Variation in temperature sum requirement for flushing of beech provenances. Silvae Genet 44:343–347
Vornam B, Gailing O, Derory J, Plomion C, Kremer A, Finkeldey R (2011) Characterisation and natural variation of a dehydrin gene in Quercus petraea (Matt.) Liebl. Plant Biol 13:881–887
Wachowiak W, Balk PA, Savolainen O (2009) Search for nucleotide diversity patterns of local adaptation in dehydrins and other cold-related candidate genes in Scots pine (Pinus sylvestris L.). Tree Genet Genomes 5:117–132
Welling A, Rinne P, Viherä-Aarnio A, Kontunen-Soppela S, Heino P, Palva ET (2004) Photoperiod and temperature differentially regulate the expression of two dehydrin genes during overwintering of birch (Betula pubescens Ehrh.). J Exp Bot 55:507–516
Williams MI, Dumroese RK (2013) Preparing for climate change: forestry and assisted migration. J For 111:287–297
Acknowledgements
The provenance experiment has been established through the realization of the project European Network for the Evaluation of the Genetic Resources of Beech for Appropriate Use in Sustainable Forestry Management (AIR3-CT94-2091) under the coordination of H.-J. Muhs and G. von Wühlisch. The experimental plot Tále was established by L. Paule. The study was supported by research grants of the Slovak Research and Development Agency APVV-0135-12 (DKr, MH, ĽD, JK, DKu, DG), APVV-0436-10 (ĽD, GJ), and Slovak Grant Agency for Science VEGA-2/0034/14 (ĽD, GJ). Technical assistance of G. Baloghová is greatly appreciated. We also thank to K. Willingham for linguistic correction.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Krajmerová, D., Hrivnák, M., Ditmarová, Ľ. et al. Nucleotide polymorphisms associated with climate, phenology and physiological traits in European beech (Fagus sylvatica L.). New Forests 48, 463–477 (2017). https://doi.org/10.1007/s11056-017-9573-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11056-017-9573-9