Abstract
Assuming a study region in which each cell has associated with it an N-dimensional vector of values corresponding to N predictor variables, one means of predicting the potential of some cell to host mineralization is to estimate, on the basis of historical data, a probability density function that describes the distribution of vectors for cells known to contain deposits. This density estimate can then be employed to predict the mineralization likelihood of other cells in the study region. However, owing to the curse of dimensionality, estimating densities in high-dimensional input spaces is exceedingly difficult, and conventional statistical approaches often break down. This article describes an alternative approach to estimating densities. Inspired by recent work in the area of similarity-based learning, in which input takes the form of a matrix of pairwise similarities between training points, we show how the density of a set of mineralized training examples can be estimated from a graphical representation of those examples using the notion of eigenvector graph centrality. We also show how the likelihood for a test example can be estimated from these data without having to construct a new graph. Application of the technique to the prediction of gold deposits based on 16 predictor variables shows that its predictive performance far exceeds that of conventional density estimation methods, and is slightly better than the performance of a discriminative approach based on multilayer perceptron neural networks.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Mineral prospectivity mapping can be viewed as a process of combining a set of input maps, each representing a distinct geo-scientific variable, into a single map depicting potential to host mineral deposits of a particular type (Bonham-Carter 1994). Assuming that we are provided with N such input maps overlaid such that each grid element (or cell) can be described by a vector x i = (x i1, x i2, …, x iN ), where x ij is the value of the jth input variable for the ith cell, the problem is to discover a mapping function f(x), output of which represents a measure of the mineralization potential for that cell. In this article, we take a data-driven approach, and therefore assume that a subset of cells is known from historical records or otherwise to contain one or more occurrences of the sought-after mineral. We also assume that mineral potential is interpreted as a probability, in which case the mapping function is onto [0, 1].
There are two general approaches to discovering such a mapping function: (i) generative approaches, and (ii) discriminative approaches. Generative approaches are based on explicitly estimating the class-conditional probability density function p(x|D) (i.e., the pdf for cells known to contain a deposit). This itself can be used as an indication of the mineralization likelihood at some test point; alternatively, p(x|D) can be combined under Bayes’ theorem with an estimate of the unconditional pdf p(x) (i.e., the pdf for all cells) to produce an estimate of the probability (Duda et al. 2001). In contrast to generative approaches, discriminative approaches, which include techniques such as logistic regression (Hosmer and Lemeshow 2000) and Multilayer Perceptron (MLP) neural networks (Bishop 1995) attempt to estimate the probabilities directly, without explicitly estimating densities. In this article we focus primarily on generative approaches.
A number of standard techniques can be applied to estimating densities. The simplest is the parametric approach, by which the form of the distribution (e.g., Gaussian) is assumed, and the problem is to estimate the values of the parameters for that distribution (e.g., mean and covariance). However, the problem with parametric approaches is that many datasets do not follow a standard distribution, and attempting to model them in this way will lead to poor estimates of the density. A more flexible alternative is to use a semi-parametric—or mixture model—approach, by which the density is modeled as a mixture of K Gaussians (Titterington et al. 1985). The problem is then to estimate the model parameters (i.e., means, covariances, and mixing coefficients for each of the K components), and this can be done using Expectation Maximization (EM) (Dempster et al. 1977). However, this added flexibility comes at a high cost, since the escalation in the number of parameters often leads to severe problems with overfitting, as well as high sensitivity to initialization. A third approach—the kernel, or Parzen, method—is a non-parametric approach that involves modeling the distribution using a series of probability windows (usually Gaussian) centered at each sample (Parzen 1962). In this case, the overall density is the average of all of the individual distributions centered at each point, and the main problem is to select an appropriate value for the smoothing parameter σ that defines the width of the windowing function. This value can often be determined using cross-validation. The kernel method has been used widely in the mineralization prediction domain (Harris and Pan 1999; Singer and Kouda 1999; Harris et al. 2003; Emilson and Carlos 2009; Wang et al. 2010), where it is often referred to as a probabilistic neural network (PNN) model. However, probabilistic neural networks (Specht 1990) are essentially just a parallelization of the general kernel-based approach, phrased in the language of neural networks, and, in this article, we prefer to use the original statistical terminology.
While the above density estimation techniques can often be applied successfully in low-dimensional input spaces, estimating densities in high-dimensional spaces is notoriously difficult (Bishop 1995), and is a direct result of the curse of dimensionality (Bellman 1961). One of the manifestations of this curse is that the number of points required to estimate a density increases exponentially with the size of the input space. This poses a particular concern in the mineralization domain, since mineralization is a rare event, and the input space will indeed be very sparsely populated. Another manifestation of the curse of dimensionality is that as the number of dimensions increases, data points are progressively concentrated more toward the boundaries of the input space; i.e., the majority of the probability mass is concentrated at the edges, not at the center (Bishop 1995; Hastie et al. 2001). This means that parametric and mixture model approaches will almost certainly break down on account of the fact that these models are parameterized by the means of the mixture components, and these means will be located far from the majority of the probability mass. Kernel methods are also likely to suffer as a result of this boundary concentration.
In this article, we describe an alternative approach to estimating densities, inspired by a recent study in the area of similarity-based learning (Bicego et al. 2006). The basic idea behind similarity-based learning is that, rather than operating on attribute data (where input takes the form of a rectangular table in which rows correspond to data points and columns to attributes), similarity-based learners operate directly on pairwise similarities between data points, usually presented in the form of a square matrix A = {a ij }, where a ij is the similarity between the ith and jth data point. There are various reasons why one might take a similarity-based approach. On some domains, there may simply be no alternative; for example, data may naturally be expressed in terms of pairwise similarities, and it may not even be possible to represent the data in any continuous metric space. On other domains, similarity-based learning may lead to better performance than methods based directly on attribute data. Indeed, we demonstrate in this article that when applied to high-dimensional geoscientific data, similarity-based density estimation leads to far better estimates of mineralization potential than the methods described above.
The article is structured as follows. We first describe how the density of a set of training points can be estimated from a graphical representation in which nodes represent training points and edge weights represent pairwise similarities between connected nodes. Importantly, we show how the density of test points can be estimated directly from such a graph (i.e., without having to construct a new graph containing test points). We then apply the method to predicting gold mineralization potential over the Ballarat region of Victoria, Australia, on the basis of 16 geoscientific predictor variables. For illustrative purposes, we first apply the technique to a two-dimensional (2D) principal component subspace of the original 16D input space and show that while parametric and mixture model approaches are inadequate even in this reduced space, and that the similarity-based method yields similar density estimates to that of a kernel-based approach. We then apply the technique to the full 16D input space and show that, in regard to its ability to predict holdout deposits, its performance far exceeds that of the kernel approach, and is slightly better than a discriminative approach based on MLPs.
Similarity-Based Density Estimation
In this section, we describe the concept of eigenvector graph centrality, and how this can be used as a measure of the likelihood of an example belonging to the distribution of training data points represented by a graph. We then describe how likelihoods can be estimated for test examples, without the need of generating a new graph and re-computing eigenvectors. Finally, we show how pairwise similarities can be calculated from attribute-based data, and discuss some of the issues associated with this.
Eigenvector Graph Centrality
Consider a graph G = (V, E), where V = {v i , i = 1, 2, …, n} is a set of vertices, each corresponding to some object in the domain, and E = {e ij } is a set of edges connecting vertices (v i , v j ). We assume that the edges are weighted with a continuous value w ij on the interval [0, 1], and that w ij represents a measure of the similarity between the objects corresponding to nodes v i and v j . Eigenvector centrality is based on the following recursive definition: a vertex is central to a graph if it is similar to other vertices which are central. While this idea is surprisingly simple, Eigenvector centrality provides a very powerful measure of the importance of a node within a graph: it forms the basis of the well-known PageRank algorithm used for ranking web pages (Brin and Page 1998), and also forms the basis for Spectral Clustering (Luxburg 2007), a family of clustering algorithms which have become very popular over the last decade.
We can capture the above definition mathematically by expressing C(v i ), the eigenvector centrality score for vertex v i , as the weighted sum of the centrality scores for all nodes to which it is connected; i.e.,
where λ is a proportionality constant. Assuming that similarities are supplied in the form of a square matrix W = {w ij }, this can more conveniently be written as the eigenvector equation
where C = (C(v 1), C(v 2), …, C(v n )) is the vector of centrality scores for vertices 1 to n.
In general, Equation (2) will have a number of eigenvectors, and some of these will have negative entries. However, from the Perron–Frobenius theorem (Grimmett and Stirzaker 2001) the dominant eigenvector of W (i.e., the eigenvector corresponding to the largest eigenvalue) will have all non-negative components, thus satisfying the requirement that centrality scores be non-negative. Note also that the dominant eigenvector will not be unique, since any linear scaling of the eigenvector will also satisfy the eigenvector equation. This means that it is relative—not absolute—centrality scores which are important. Without any loss of generality we will assume that the dominant eigenvector has been normalized such that its components sum to unity.
In principle, any eigenvalue algorithm can be used to find the dominant eigenvector. A general and robust approach is power iteration, which begins with a random vector C 0, and simply iterates the step \( {\mathbf{C}}_{k + 1} = {\mathbf{WC}}_{k} \) until convergence, when C will be the dominant eigenvector. Algorithms based on matrix decomposition techniques can also be applied, and avoid the need for iteration; however, these may fail because of bad scaling unless the similarity matrix is appropriately normalized. A normalization technique commonly applied in the spectral clustering literature (Luxburg 2007) is to construct a symmetric similarity matrix, S, known as the graph Laplacian, and defined as \( {\mathbf{S}} = {\mathbf{D}}^{ - 1/2} {\mathbf{WD}}^{ - 1/2} \) where \( D_{ii} = \sum\nolimits_{j = 1}^{N} {w_{ij} ,\;i = 1, \ldots ,n} \). All the results presented in this article are based on graph Laplacians.
Estimating Likelihoods on Test Data
Each entry of the dominant eigenvector of S represents a relative measure of the centrality of the corresponding training example, and we assume that these entries are proportional to likelihoods. One method of determining the centrality of a test example is to insert it into the graph and re-compute the eigenvector. However, apart from the additional computational expense incurred, this will perturb the centrality values of the training examples, and is especially a problem if the number of training examples is small.
A better approach is to estimate the centrality of a test example directly from the dominant eigenvector of the original graph. It follows from Equation (2) that the ith value of the eigenvector is equal to its dot product with the ith row of S; i.e., \( C(v_{i} ) = \sum\nolimits_{j = 1}^{N} {s_{ij} C(v_{i} )} \), where s ij are the components of S. Thus, if we can estimate a vector s t = (s t1, s t2, …, s tn ), where s tn is the similarity between the test example and the nth training example, then the dot product of s t and the principal eigenvector will provide an estimate of the centrality (i.e., likelihood) of the test example.
In order that the centrality score assigned to the test example is consistent with those for labeled examples, we must ensure that similarity values for the test example are normalized in a manner consistent with the use of the graph Laplacian. Defining the graph Laplacian as above means that entries of S and W are related according to \( s_{ij} = {{w_{ij} } \mathord{\left/ {\vphantom {{w_{ij} } {\sqrt {\sum\nolimits_{i = 1}^{N} {w_{ij} } \sum\nolimits_{j = 1}^{N} {w_{ij} } } }}} \right. \kern-\nulldelimiterspace} {\sqrt {\sum\nolimits_{i = 1}^{N} {w_{ij} } \sum\nolimits_{j = 1}^{N} {w_{ij} } } }} \). It follows that the components of the normalized similarity vector for the test example vector s t are given by \( s_{{{\text{t}}i}} = {{w_{{{\text{t}}i}} } \mathord{\left/ {\vphantom {{w_{{{\text{t}}i}} } {\sqrt {\sum\nolimits_{i = 1}^{N} {w_{{{\text{t}}i}} } \sum\nolimits_{j = 1}^{N} {w_{ij} } } }}} \right. \kern-\nulldelimiterspace} {\sqrt {\sum\nolimits_{i = 1}^{N} {w_{{{\text{t}}i}} } \sum\nolimits_{j = 1}^{N} {w_{ij} } } }} \), where w ti is the similarity between the test example and the ith training example. Finally, the eigenvector centrality of the test example can be expressed as \( C(t) = \sum\nolimits_{i = 1}^{N} {s_{{{\text{t}}i}} C(v_{i} )} \).
Converting Distances to Similarities
The above discussion has assumed an input matrix W containing pairwise similarities. In many cases, pairwise similarities may not be directly available, and may need to be computed from the given data. We assume here that attribute data are supplied; that these data represent points in a Euclidean space, and that similarities are calculated by passing Euclidean pairwise distances through the monotonically decreasing function \( f(x) = \exp ( - x^{2} /2\sigma^{2} ) \), where x is the Euclidean distance, and σ controls the rate at which similarity falls off with distance. Clearly, the final distribution of centrality values for the nodes on the resulting graph will depend on σ: if similarities fall off quickly with distance (i.e., small σ), then the density may be more sharply peaked around either individual data points or closely clustered collections of points, resulting in overfitting to the training data; conversely, if similarities fall of slowly with distance (high σ), then the overall density will be smoother, and may result in underfitting of the training data. How might an appropriate value for σ be determined?
A common means of optimizing parameters when estimating densities using Gaussians is to perform cross-validation on the training data, and to select the parameter values that maximize the likelihood (or equivalently, that minimize the negative log likelihood) on holdout examples. This is possible in the Gaussian case, since Gaussians are normalized to unit area, and hence so too can a sum of Gaussians be normalized. However, in the case of similarity-based density estimation, the components of the dominant eigenvector represent only a relative measure of centrality. This means that determining parameter values through cross-validation is no longer generally possible, and that we must therefore resort to using some other heuristics to determine these parameters. For the case of predicting mineralization likelihood, we provide some heuristics in the next section. We note also that that even for kernel density estimation, determining the optimal kernel width through cross-validation on high-dimensional spaces can be very unreliable, as will also be demonstrated later.
Empirical Results
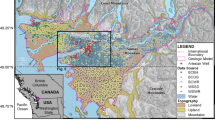
This section reports on the application of the technique described above to predicting gold mineralization in the vicinity of Castlemaine, located in the Southeastern region of Victoria, Australia.
Southeastern Victoria was the site of extensive gold mining in the nineteenth Century. Almost half of the gold found occurred in primary deposits, particularly quartz veins or reefs, in which it was deposited in cracks that opened up during the faulting and folding of Palaeozoic sandstone and mudstone beds between 440 and 360 million years ago. The remainder has been found in secondary (alluvial) deposits in soil and creek beds. The general region is well explored, and any remaining deposits are expected to be hidden under basalt cover (Lisitsin et al. 2007). Information on Victorian geology can be found in Cochrane et al. (1995) and Clark and Cook (1988). The Castlemaine Goldfield is described in Willman (1995).
The specific area selected for this study extends from a Northwest corner with coordinates 251,250 mE, 5,895,250 mN, to a Southeast corner with coordinates 258,250 mE, 5,885,000 mN, where all specified coordinates are Northings/Eastings referenced according to AMG Zone 55 AGD 66. This selection was based the range of data types available (geophysical, geochemical and geological), and the coverage of that data. Magnetic and radiometric data have full coverage over the region, and the density of geochemistry sampling points is sufficient to provide an acceptable interpolated coverage over most of the area. There are also a number of known fault zones in the region. The number of documented reef gold deposits is 148. Additional gold deposit locations have also been recorded, but the historical information on these does not indicate either the type of occurrence (i.e. reef or alluvial) or their significance. In this study we use only the 148 documented reef deposits.
Input data consists of 16 input layers: three based on magnetics (magnetic field intensity, first derivative of magnetic field intensity, and automatic gain control filtered magnetics); five layers based on radiometrics (Th, U, K, TotalCount, K/Th); seven based on geochemistry (Au, As, Cu, Mo, Pb, W, Zn), and distance to closest fault. Each input variable was normalized by subtracting the mean and dividing by the standard deviation. The study region was represented by a rectangular grid of 141 × 206 cells with resolution 50 m × 50 m.
We note that there may be some overlap between some of the input variables described above. For example, because magnetic first derivative maps out the structures, it is expected to be correlated with the distance to the closest fault layer. The layers will not, however, be identical, since the magnetics may indicate structure additional to the known faults, and the general philosophy we adopt is that any data that are considered both relevant and non-redundant should be incorporated. In any case, while the presence of duplicate or highly correlated layers may overemphasize the importance of that layer, particularly when the number of input dimensions is small, the degree of overemphasis decreases as the dimensionality of the input space is increased. The question of the relative importance of inputs is a matter that relates to virtually all pattern recognition tasks, and in the context of kernel density estimation, it can be controlled through the use of different sigmas; i.e., using a large sigma value for a particular input will de-emphasize the importance of that variable relative to inputs with smaller sigmas. In the context of the similarity based method, it can be controlled through replacing Euclidean distance with Mahalanobis distance. This is discussed further in the final section of the article.
We also note that an important consideration in choosing the study region described above is that it does not contain basalt cover. There are two reasons for this. First, the presence of basalt leads to significantly different characteristics in geophysical input signals compared to those obtained in the absence of basalt, and attempting to simultaneously discover predictive patterns for both covered and uncovered regions would yield poor results because of the confounding of these characteristically different signals. Second, historical deposit information is only generally available for regions not under cover. Owing to the extensive exploration activities conducted over the region it is highly unlikely that undiscovered deposits exist in areas not under basalt cover. However the wealth of data available for the region (i.e., high-dimensional multi-source input data and sufficient historical deposit information) makes it a very suitable domain for comparing the proposed method with other predictive methods.
A Two-Dimensional Illustrative Example
For demonstrative purposes, so that we can visualize the estimated densities, we first consider a 2D input space consisting of the first two principal components of the full 16D dataset. Principal Component analysis (Pearson 1901; Jolliffe 2002) is a dimensionality reduction method that operates by transforming the original data into a new orthogonal coordinate system in such a way that the first principal component accounts for as much of the variability in the data as possible, with succeeding components accounting for as much of the remaining variability as possible. We first apply parametric and semi-parametric approaches and show that these can be problematic even in two dimensions. We then compare the similarity-based and kernel approaches.
Gaussian Mixture Models
As stated in the Introduction, mixture models are a semi-parametric approach in which the density is modeled as a linear combination of C Gaussian component densities \( p({\mathbf{x}}|m) \) in the form \( \sum {\pi_{m} p({\mathbf{x}}|m)} \), where the \( \pi_{{m}} \) are called mixing coefficients, and represent the prior probability of data point x having been generated from component m of the mixture. The problem is to determine these means, covariances, and mixing coefficients, and this can be done using the Expectation–Maximization (EM) algorithm (Dempster et al. 1977), in which an Expectation step (E-step), and followed by a Maximization step (M-step), are iterated until convergence. The E-step computes the cluster membership probabilities:
where \( \hat{\varvec{\upmu }}_{m} \) and \( \hat{\varvec{\Upsigma }}_{k} \) are the current estimates of the mean and covariance of component m. In the M-step, these probabilities are then used to re-estimate the parameters:
Figure 1 shows the density contours resulting from density estimation based on mixtures of one or more Gaussians. Horizontal and vertical axes represent first and second principal components, respectively. Points represent 25 randomly selected mineralized cells from which the densities are estimated (i.e., training points), and crosses represent the remaining 123 mineralized cells, which are treated as holdout examples, and can be employed to assess the quality of the density estimate (i.e., its ability to generalize to the prediction of holdout points).
Figure 1a models the density using a single Gaussian. As stated above, using a single Gaussian often fails to adequately capture the structure in the data, and this is clearly apparent in this case, in which visually the data can be seen to contain a cluster of points centered at approximately (0, 1), and another cluster centered in the vicinity of (3, −1). While the use of two Gaussians, as shown in Figure 1b, better captures the structure in the training data, it does not generalize well to predicting the hold-out data, as evidenced by the dense concentration of holdout points in the bottom-left and top-right regions, where predicted likelihood is low. Figure 1c and d, each of which models the training data using three Gaussians, but with a different EM initialization in each case, likewise performs poorly in generalizing to the prediction of holdout data, and also demonstrates the high sensitivity of mixture model approaches to initialization. Clearly, there are difficulties in applying mixture model approaches to even 2D data, and the curse of dimensionality means that these difficulties will only be exacerbated in higher dimensional spaces.
Similarity-Based and Kernel Approaches
The kernel approach estimates densities as the weighted sum of Gaussian kernels centered at each training point. The density p(x) can be expressed as,
where \( \varvec{\Upsigma} \) is the covariance of the Gaussian kernel, and N is the number of training points. The covariance matrix \( \varvec{\Upsigma} \) can be selected by taking into account the fact that the variance in the data may differ across variables; however, since we are using (normalized) Principal Components, we use a spherical covariance matrix, in which case, the covariance can be replaced by the single variance parameter σ 2k , where the subscript k (for kernel) is used for avoiding confusion with the sigma used in the distance-to-similarity conversion discussed earlier. The optimal value for σk can be determined through leave-one-out cross validation on the training data; i.e., by estimating the likelihood at each point on the basis of the other training points, and selecting the value of σk that minimizes the overall negative log likelihood. Negative log likelihoods for a range of σk values are shown in Figure 2a, from which it can be seen that optimal value is approximately 0.75. Figure 2b shows contours of the corresponding density estimate.
Figure 2c shows the density estimated using the similarity-based approach. The only parameter involved in this case is σsb, which is the parameter controlling the rate at which similarity falls off with distance in the distance-to-similarity conversion. Because densities estimated by the similarity-based approach cannot be normalized to unit area we cannot use cross-validation to determine σsb, and in this case we have used the same value as for the kernel approach (i.e., σsb = σk = 0.75). (In the next section, we show that the kernel approach is actually equivalent to a graph-centrality density estimation based on a simpler measure of similarity, and that, at least in the case of low-dimensionality input spaces, optimal σ values for the similarity-based and kernel approaches are likely to be similar).
The general shape of the contours in Figure 2b and c are similar, although the contours in Figure 2c are spread further over the input space than are those in Figure 2b, which tends to be more sharply peaked. Visually, they appear to both perform far better than any of the methods of Figure 1 both in modeling the training data, and also in generalizing to hold-out examples. In the next sub-section, we compare the similarity-based and kernel methods over the full 16D input space.
Full 16-Dimensional Input Space
The only parameter involved in the similarity-based method is σsb, and as described above, it is not possible to use cross-validation to determine this parameter. However, some indication of the appropriateness of various σsb values can be gained by considering the histogram of likelihoods resulting from estimating the density over all cells in the study region. For small values of σsb, the distribution will be peaked around the training points, and means that we should expect to see high likelihoods assigned to very few points, and low values assigned to the majority. Conversely, when σsb is large, the distribution will be relatively flat over the input space, and means that all cells would be assigned likelihoods distributed within a narrow range about the mean likelihood.
Figure 3 shows the histograms of likelihoods corresponding to several σsb values, and calculated based on all 148 training points. In order that histograms can be meaningfully compared, the likelihoods in each case have been scaled to sum to 148 over all cells in the study region. Scaling in this way is useful, because it means that when interpreted as probabilities, the values can directly be compared with the prior probability of mineralization value of 0.0051 (i.e., 148/29046, based on the number of known mineralized cells as a fraction of the total number of cells). As can be seen from the histograms, as σsb is increased, the distributions do indeed progressively become centered more narrowly around the mean.
Examination of the histograms reveals a convenient heuristic which we can use to help select an appropriate σsb value. Mineralization is a rare event, and on the basis of expert knowledge, we would probably rule out the possibility that a cell has, say, a probability of 20% of containing a deposit, such a value being considered too high to be realistic. Similar arguments apply to small probabilities, and we may be unlikely to accept, for example, that 90% of cells have a mineralization probability of less than 0.005%. Based on these types of arguments and our knowledge of the study area, we believe that the distributions in the histograms of Figure 3b and c provide realistic estimates of the range and distribution of values that we would expect in this domain. We note, however, that these histograms display only the overall distribution of likelihoods, and provide absolutely no indication of how accurately these likelihoods reflect the holdout deposits. Before examining their predictive performance, we look at the histograms for the kernel approach.
Figure 4a shows how the negative log likelihood obtained using leave-one-out cross-validation on all 148 training examples varies with σk, and Figure 4b shows the histogram corresponding to σk = 0.45, for which the negative log likelihood is a minimum. As seen from the histogram, some cells are assigned mineralization probabilities of 44%, which is clearly unrealistic, suggesting that there has been a breakdown in cross-validation procedure. Training points close to some test point contribute more to the density estimate at that test point than training points further away, and the low σk value of 0.45 suggests that the overall likelihood is being dominated by a few closely neighboring points—another manifestation of the curse of dimensionality. Higher values for σk yield much more realistic results, and as can be seen from Figure 5, a value of 1.6 results in a distribution very close to those in Figure 3b and c, obtained using similarity-based methods.
A useful means of comparing the predictive performance of the methods is to plot cumulative deposits versus cumulative area curves. These curves can be constructed by ranking cells according to their assigned mineralization probability value, and plotting the cumulative deposits against cumulative area as the posterior probability is decreased from its maximum to its minimum value. The area under such a curve provides a measure of the predictive performance of the technique.
Figure 6a and b shows the cumulative deposits versus cumulative area curves corresponding respectively to the similarity-based and kernel approaches. (Note that the cumulative deposits and cumulative area are expressed as a percentage of the total.) Solid lines represent the actual predictive performance; dashed lines show the cumulative sum of probabilities. The proximity of the solid and dashed curves provides an indication of how well-calibrated the probabilities are. The results in each case are based on leave-one-out testing; i.e., 148 models were estimated, each using a different subset of 147 mineralized examples to develop a model. These models were then used to estimate the probability of the corresponding hold-out example. Probabilities for the non-mineralized cells were averaged over the 148 models. The curves therefore represent a true test of the ability of a technique to predict mineralization on unseen data.
While the area under a curve represents a measure of the overall predictive performance, we are typically more interested in how well the technique works in predicting mineralization in the areas predicted as most favorable; e.g., the percentage of deposits appearing in the top 5% of cells. Table 1 shows summary data for the similarity-based and kernel method. For comparative purposes, we also show results from applying discriminative density estimation using MLPs. Details on applying MLPs can be found in Skabar (2005, 2007).
As can be seen from the table, the similarity-based and MLP approaches yield similar results to each other, and markedly outperform the kernel approach in their ability to correctly predict the presence of deposits in regions predicted most likely to contain mineralization, with the proportion of deposits predicted in both the upper 5% and upper 10% regions being approximately double the proportion predicted by the kernel approach. It is not surprising that the MLP approach achieves such results, since discriminative approaches are generally regarded as preferable to density estimation based approaches in high dimensional input spaces (Bishop 1995). The difficulty with MLPs, however, is that there are many factors that affect their performance, including network architecture (e.g., number of hidden layer units), weight optimization algorithm, early stopping point for training, and regularization coefficient values. Selecting appropriate combinations of values for these parameters can be very difficult, and usually requires a complex cross-validation procedure. In contrast, the similarity-based method requires only a single parameter to be determined; i.e., the value of σsb used in the distance-to-similarity conversion. While we cannot directly use cross-validation to determine σsb, the results have shown that the heuristic described above has led to an appropriate choice for this parameter.
Discussion and Concluding Remarks
While the similarity-based and kernel approaches yield similar results when the dimensionality of the input space is low, the similarity-based approach achieves markedly superior performance in higher-dimensional input spaces. To explain this phenomenon, it is insightful to recognize that the kernel approach can in fact be considered equivalent to similarity-based density estimation using degree—as opposed to eigenvector—centrality. While eigenvector centrality computes centrality recursively, the degree centrality, C D(v), of a vertex v in some graphs is defined more simply as the sum of the weights of the edges incident on it; i.e., \( C_{\text{D}} (v) \propto \sum\nolimits_{j = 1}^{N} {w_{ij} } \). If these (similarity) weights are derived from Euclidean distances according to \( w_{ij} = \exp ( - x_{ij}^{2} /2\sigma^{2} ) \), where x ij is the Euclidean distance between points i and j, then the degree centrality calculated at some test point is effectively just the weighted sum of Gaussians centered at the training points and evaluated at the test point, and this is exactly what is calculated by the (Gaussian) kernel approach. While in the case of degree centrality the density estimate at some point is based only on N distances (i.e., the distances between the test point and each of the training points), in the case of eigenvector centrality, the estimate is based on N + N(N − 1)/2 distances; i.e., the N distances between the test and training points, and (implicitly) on each of the pairwise distances between training points. It is because the similarity-based method used in conjunction with eigenvector centrality utilizes this richer information set that it is better able to estimate densities in higher dimensional spaces.
The kernel-based method used in this article was based on the use of a common sigma value for each input variable, and performance can sometimes be improved by allowing separate sigmas for each variable. The difficulty with this, however, is that using separate sigmas for each of the 16 input variables increases the number of model parameters which must be estimated from 1 to 16. Estimating these parameters in a 16D input space using only 148 training points would result in exceedingly poor estimates of the sigmas, and would almost certainly result in predictive performance inferior to that obtained using a single sigma. However, just as the kernel method can be extended to allow sigmas for each variable, so too can the similarity-based method be extended in an analogous way by simply replacing Euclidean distance with Mahalanobis distance as the measure of separation between data points. Just as the use of multiple sigmas with the kernel method may lead to improved performance (given sufficient data to reliably estimate the sigmas), so too might the use of Mahalanobis distance be expected to improve on the use of Euclidean distance.
Although we have applied similarity-based density estimation to (Euclidean) attribute data, it can be applied to any domain in which pairwise similarities are available. Indeed, a very attractive feature of the technique is that when applied directly to similarities, it is absolutely parameter free. Its ability to estimate densities in non-metric spaces, where distances need not satisfy the triangle rule, make it useful for domains in which similarities between objects are expressed using some form of human judgment. A mineral geoscientist may, for example, use his/her knowledge and expertise to assign a similarity between two locations that may be very difficult to quantify in terms of available attribute-based data.
One of the limitations of the technique is its memory requirements. The full matrix of pairwise similarities must be kept in memory, and this could be expensive if the number of training points is high. This is not so much of a problem in the mineral prediction domain, since the number of known deposits will normally not be high.
While we have concentrated in this article on the use of similarity-based methods to estimate densities, the technique can also be extended to perform classification. This could be done by modeling each of the classes as a separate graph, estimating the corresponding class-conditional densities, and classifying a test example into the class for which the density, scaled by the prior is greatest. An example might be land-cover classification.
References
Bellman, R. E. (1961). Adaptive control processes. Princeton, NJ: Princeton University Press.
Bicego, M., Murino, V., Pelillo, M., & Torsello, A. (2006). Similarity-based pattern recognition. Pattern Recognition, 39(10), 1813–1814.
Bishop, C. (1995). Neural networks for pattern recognition. Oxford: Oxford University Press.
Bonham-Carter, G. F. (1994). Geographic information systems for geoscientists: Modeling with GIS. Oxford: Pergamom Press.
Brin, S., & Page, L. (1998). The anatomy of a large-scale hypertextual web search engine. Computer Networks and ISDN Systems, 30(1–7), 107–117.
Clark, I., & Cook, B. (Eds.). (1988). Victorian geology excursion guide. Canberra: Australian Academy of Science.
Cochrane, G. W., Quick, G., & Spencer-Jones, D. (1995). Introducing victorian geology (2nd ed.). Melbourne: Geological Society of Australia Incorporated (Victorian Division).
Dempster, A. P., Laird, N. M., & Rubin, D. B. (1977). Maximum likelihood from incomplete data via the EM algorithm. Journal of the Royal Statistical Society B, 39, 1–38.
Duda, R. O., Hart, P. E., & Stork, D. G. (2001). Pattern classification (2nd ed.). New York: Wiley.
Emilson, P., & Carlos, R. (2009). Probabilistic neural networks applied to mineral potential mapping for platinum group elements in the Serra Leste region, Carajas Mineral Province, Brazil. Computers and Geosciences, 35(3), 675–687.
Grimmett, G. R., & Stirzaker, D. R. (2001). Probability and random processes (3rd ed.). Oxford: Oxford University Press.
Harris, D., & Pan, G. (1999). Mineral favorability mapping: a comparison of artificial neural networks, logistic regression, and discriminant analysis. Natural Resources Research, 8(2), 93–109.
Harris, D., Zurcher, L., Stanley, M., Marlow, J., & Pan, G. (2003). A comparative analysis of favorability mappings by weights of evidence, probabilistic neural networks, discriminant analysis, and logistic regression. Natural Resources Research, 12(4), 241–256.
Hastie, T., Tibshirani, R., & Friedman, J. (2001). The elements of statistical learning: Data mining, inference, and prediction. New York: Springer.
Hosmer, D. W., & Lemeshow, S. (2000). Applied logistic regression (2nd ed.). New York: Wiley.
Jolliffe, I. T. (2002). Principal component analysis. Springer series in statistics (2nd ed.). New York: Springer.
Lisitsin, V., Olshina, A., Moore, D. H., & Willman, C. E. (2007). Assessment of undiscovered mesozonal orogenic gold endowment under cover in the northern part of the Bendigo Zone. GeoScience Victoria Gold Undercover Report 2. Department of Primary Industries.
Luxburg, U. V. (2007). A tutorial on spectral clustering. Statistics and Computing, 17(4), 395–416.
Parzen, E. (1962). On estimation of a probability density function and mode. Annals Mathematical Statistics, 33(3), 1065–1076.
Pearson, K. (1901). On lines and planes of closest fit to systems of points in space. Philosophical Magazine, 2(6), 559–572.
Singer, D. A., & Kouda, R. (1999). A comparison of the weights-of-evidence method and probabilistic neural networks. Natural Resources Research, 8(4), 287–298.
Skabar, A. (2005). Mapping mineralization probabilities using multilayer perceptrons. Natural Resources Research, 14(2), 109–123.
Skabar, A. (2007). Mineral potential mapping using Bayesian learning for multilayer perceptrons. Mathematical Geology, 39(2), 439–451.
Specht, D. F. (1990). Probabilistic neural networks. Neural Networks, 3(1), 109–118.
Titterington, D. M., Smith, A. F. M., & Makov, U. E. (1985). Statistical analysis of finite mixture distributions. New York: Wiley.
Wang, G., Zhang, S., Yan, C., & Song, Y. (2010). Probabilistic neural networks and fractal method applied to mineral potential mapping in Luanchuan region, Henan Province, China. In Proceedings of sixth international conference on natural computation (Vol. 2, pp. 1003–1007).
Willman, C. E. (1995). Castlemaine goldfield: Castlemaine-Chewton, Fryers Creek 1:10000 maps geological report, Geological Survey report 106. Energy and Minerals Victoria.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Skabar, A.A. Mineral Prospectivity Prediction from High-Dimensional Geoscientific Data Using a Similarity-Based Density Estimation Model. Nat Resour Res 20, 143–155 (2011). https://doi.org/10.1007/s11053-011-9145-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11053-011-9145-1