Abstract
Lung cancer is the leading cause of death among cancer-related death. Like other cancers, the finest solution for lung cancer diagnosis and treatment is early screening. Automatic CAD system of lung cancer screening from Computed Tomography scan mainly involves two steps: detect all suspicious pulmonary nodules and evaluate the malignancy of the nodules. Recently, there are many works about the first step, but rare about the second step. Since the presence of pulmonary nodules does not absolutely specify cancer, the morphology of nodules such as shape, size, and contextual information has a sophisticated relationship with cancer, the screening of lung cancer needs a careful investigation on each suspicious nodule and integration of information of all nodules. We propose deep CNN architecture which differs from those traditionally used in computer vision to solve this problem. First, the suspicious nodules are generated with the modified version of U-Net and then the generated nodules become an input data for our model. The proposed model is a multi-path CNN which exploits both local features as well as more global contextual features simultaneously to automatically detect lung cancer. To this end, the model used three paths, each path employed different receptive field size which helps to model distant dependencies (short and long-range dependencies of the neighboring pixels). Then, to further upgrade our model performance, we concatenate features from the three paths. This balance the receptive field size effect and makes our model more adaptable to the variability of shape, size, and contextual information among nodules. Finally, we also introduce a retraining phase system that permits us to tackle difficulties related to the imbalance of image labels. Experimental results on Kaggle Data Science Bowl 2017 challenge shows that our model is better adaptable to the described inconsistency among nodules size and shape, and also obtained better detection results compared to the recently published state of the art methods.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Worldwide, the top cause of death among cancer related death is lung cancer (Rebecca et al. 2018; WHO 2018). According to the World Health Organization (WHO) report 2018, lung cancer is responsible for an estimated 1.76 million deaths (WHO 2018). Only in the United States of America, among the cancer-related deaths, 83,550 deaths were estimated because of lung cancer, in 2018 (Rebecca et al. 2018). This number is expected to be higher in developing countries. Figure 1 shows the leading cancer types for the estimated deaths by sex in the United States of America, where lung cancer is leading in both sexes. Like other cancers, the finest solution for lung cancer is early diagnosis and treatment. To this end, the primary and critical step for early diagnosis and treatment of lung cancer is identifying the lung whether it is infected by cancer or not, with better screening approaches leading to polished patient result. The national lung screening trial (NLST) determined that screening with low dose helical Computed Tomography (CT) scan decreased death rate by 20% contested to single view radiography in high-risk demographics (The National Lung Screening Trial Research Team 2011). However, screening for lung cancer is prevalent to false positive, increasing cost, and causing tension for patients (Patz et al. 2014). Computer-aided diagnosis (CAD) of lung cancer provides an increased attention in early screening and a decreased false positive rate in diagnosis.
Leading Cancer types for the estimated deaths by sex in United States, 2018 (Rebecca et al. 2018)
After its great achievement in natural image recognition and classification, CAD of medical imaging using deep learning with CNN methods has achieved great success over the other state of the art for automated medical imaging applications (Alvarez et al. 2012), and are for example able to detect skin cancer metastases (Liu et al. 2017), obtaining considerably better sensitivity performance than human. Also, Kingsley et al. 2017 addressed automatic lung cancer screening using 3D-CNN, and obtained promising performance, with some drawbacks such as speed inference and memory efficiency.
Because of the various reasons mentioned in this paper, however, the existing automatic screening of lung cancer methods have not demonstrated sufficiently accurate and robust result for clinical use. Some of the key challenges of lung cancer detection are
-
The complexity of contextual information among nodules The nodule is a solid clumps of tissue that exists in and around the lungs. These nodules are visible in CT scan images and have complex contextual information, they can be cancer (Malignant) or non-cancer (Benign). Because of their sophisticated contextual information, the existing cancer screening algorithm faces the problem of accuracy, and typically very tough for Doctors.
-
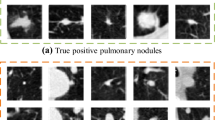
The inherent size and shape of cancer nodules The Morphology of the cancer nodules and nodules in general, are varying across the CT scan images of a lung. The inconsistency of cancer nodules shape and size will create an ambiguity on radiologist and/or oncologists during screening. Due to this, radiologists and/or oncologists recommend for further action such as monitoring, blood test, and biopsy, and as a result, it becomes a burden for patients. Not only human, but it is also not easy for an algorithm to localize the cancer properties with their respective size and shape. Figure 2 demonstrates the mentioned variation among nodules.
Cancer and non-Cancer lung nodules detection methods using CNN can be determined by how neighboring pixels are well modeled, i.e., modeling short (local) and long (global) range dependencies determine model learning accuracy. Mostly, the traditional CNN models used to detect lung cancer are utilizing 3 × 3, 5 × 5, or 7 × 7 receptive field size to model distant dependencies in a single pathway (Kingsley et al. 2017; Havaei et al. 2017; Pereira et al. 2016). Whereas these methods are considerably provided better performance than the traditional lung cancer detection methods, they lack to jointly integrate local and global range dependencies. Different receptive field sizes are not considered jointly, rather it acts on feature maps with fixed receptive field size, and due to this, modeling the neighboring pixel somehow deteriorate the model performance. On the other way, those aforementioned problems are not jointly considered, since the size and shape, and contextual information is all about how modeling neighboring pixels. For example, from Fig. 3, one can observe that the values \( g_{i1} \), \( g_{j1} \), and \( g_{k1} \) are the outputs of the receptive fields \( R_{i} \), \( R_{j} \), and \( R_{k} \) respectively, using convolution. \( g_{i1} \) approximates the receptive field \( R_{i} \), which means all pixels of \( R_{i} \) are approximated by \( g_{i1} \) using convolution. The same is true for \( g_{j1} \) and \( g_{k1} \). Accordingly, the entire feature maps are modeled. However, if we observe what \( g_{i1} \), \( g_{j1} \), and \( g_{k1} \) computes are quite different, because the receptive field size used are different. The best size depends on a task. Therefore, it is important to design an architecture which averages the influence of receptive field size.
In addition, the existing models for automatic lung cancer detection using CNN faces a shortage and class imbalance of labeled data. For example, the CT scan of Kaggle Data Science Bowl 2017 challenge dataset is highly data imbalanced, the malignancy to benign ratio is 1:7.
To address these aforementioned challenges, this paper proposes a multi-path CNN which in general we provide flexible and adaptable model used to detect medical images, and lung cancer in particular. The model is designed in such a way that those key challenges are addressed by our method. Also, compared to 3D-CNN which worth much time and memory, we focus on 2D-CNN with multipath and provided competitive performance. The proposed multi-path CNN is similar to the traditional CNN architecture, however, it differs in structure. It has three paths, each path is intended to model distant dependencies (neighboring pixels), where one can relate these distant dependencies with the pointed shape and size of cancer existing in the lung. The first and the second paths are intended to model local dependencies (more focus on shape and size), and the third path is intended to model long-range dependencies (contextual information). In general, our architecture is designed in a unified form to tackle the distant dependencies. We also address the class imbalance problem. Our work can be summarized as follows:
-
We propose a novel automatic multi-path architecture used to detect lung cancer with CNN. Our architecture is designed in such a way that various shape and size of cancer and non-cancer nodules features are learned.
-
To deal with the crucial class imbalance problem, we introduce a technical approach with retraining phase. Details of the contribution are found in Sect. 3.1.
-
We also deal with concatenation and averaging features from the three paths to polish the model performance, where we found better accuracy.
The rest of this paper is organized as follows. Section 2 briefly review related works. In Sect. 3 we present dataset used and preprocessing (segmentation of the lung, detecting the suspicious nodule and training U-Net). The detail of the proposed multi-path CNN with its detail explanations of each path is given in Sect. 4. The experimental results and discussions are well presented in Sect. 5 and finally, Sect. 6 concludes the paper.
2 Related works
Automatic lung cancer detection algorithm aids an expert in understanding medical images, granting for medical image analysis with higher sensitivity and specificity, which is very crucial for patients. As pointed by Muhammad et al. (2017), the number of publications contributed to lung cancer detection has grown rapidly in the last many years. This observation not only underlines the importance of automatic lung cancer detection tools but also reveals that research in that area is still a work in progress.
In general, there exist various image detection approaches (especially those devoted to lung cancer), and they can be categorized into those based on handcrafted models and those based on discriminative models. The handcrafted models rely heavenly on domain-specific knowledge about the existence of both non-cancer nodules and cancerous ones. The nodules appearance is challenging to characterize, and existing handcrafted models usually misscreening as cancerous or non-cancerous nodules which divert from the normal one (Clark et al. 1998; Lin and Yan 2002). On the other hand, unlike handcrafted models, discriminative methods exploit little prior knowledge on the lungs anatomy. It mostly focuses on extracting low-level features, directly modeling the relationship between these features and the label of the provided value.
A thorough review is beyond the scope of this paper, but we give here the recent discriminative models designed to detect an image. Most of these approaches are designed for 2D image detection. Faster-RCNN (Ren et al. 2015), in which some bounding boxes are proposed in the first phase and the class decision is computed in the second phase. More recent approaches have a single stage, in which the bounding boxes and class probabilities are predicted instantaneously (Redmon and Farhadi 2016) or the class probabilities are estimated for default boxes without proposal generation (Liu et al. 2016). Overall, single stage approaches are quicker but two-stage approaches are more accurate. In the case of single class object detection, the second stage in the two-stage approaches is no longer wanted and the approaches degenerate to single-stage methods.
An extension from 2D to 3D CNN of image detection is also considered. For example, considering a CT scan image of the lung, (Alvarez et al. 2012; Kingsley et al. 2017; Havaei et al. 2017) introduced 3D CNN to detect lung cancer with multi-stage CNN and have provided relatively promising performance. However, the architecture takes much time to train and also needs much memory. This is a very typical problem when lung cancer detection is to be employed in our day-to-day work. Also, these methods do not take into account those problems described earlier in this paper, but those problems are addressed in this paper.
3 Dataset and preprocessing
3.1 Dataset
To train our model, we used the Kaggle Data Science Bowl (KDSB) 2017 challenge (KDSB 2017). The dataset is provided in DICOM file format having patient Id and labels (as shown in Table 1). It comprises labeled data for 2101 patients, where a label 0 is for the patient with no cancer and 1 is for the patient with cancer. For each patient, the CT scan comprises a variable number of images (normally around 100–400, each image is a 2-D axial slice) of 512 × 512 pixels. After some preprocesses are applied to these 2D axial slice images, the model is trained with these images.
Since the Kaggle dataset alone proved to be insufficient to accurately classify the validation set, we used patient lung CT scan dataset with labeled nodules from the Lung nodule analysis 2016 (LUNA16) Challenge (LUNA 2016). The LUNA16 dataset comprises labeled data for 888 patients, where for each patient the data comprises of CT scan data and a nodule label (list of nodule center coordinates and diameter). Similar to the CT scan images of KDSB, each patient of the LUNA16 dataset comprises CT scan images of 512 × 512 pixels.
We divide the images of LUNA16 into a training set of size 710 and a validation set of size 178 to train a modified U-Net. The trained U-Net helps us to segment suspicious nodules regions of KDSB CT scan images (i.e., the U-Net is tested with KDSB dataset). Once the suspicious nodules regions are approximated by U-Net, we divide these approximated images into a training set, validation set, and test set to train our proposed architecture. Figure 6 shows the suspicious nodules samples which are segmented by U-Net.
The KDSB 2017 dataset is highly data imbalanced, where 70% of the patients are free of cancer and 30% of the patients are with cancer. Choosing patches from true distribution would cause the model to be highly influenced by healthy patches and this will affect the training accuracy of CNN models.
To circumvent this, we initially build our patches dataset such that the two labels (cancer, non-cancer) are equiprobable. To make they are equiprobable, we perform data augmentation (Krizhevsky et al. 2012) on images whose label is 1 (i.e., we augment the set of malignant nodules by filliping and 90-degree rotations) and perform training. Next, we consider the unbalanced data and retrain only the output layer (i.e., keeping the weight of all other layers fixed) with a more representative distribution of the labels. In this way, we get the best result.
3.2 Preprocessing
3.2.1 Lung segmentation
A CT scan image of a lung comprises not only the lung but also other tissues, such as bone, air, blood, and water. These substances are not important, their presence affects the ability that the model characterizes the nodules, and thus we need to exclude them.
Each CT scan of KDSB consisting of multiple 2D axial slices with pixel values in the range [− 1024, 3071] corresponding to Hounsfield Unit (HU), which is the quantitative scale for describing Radio-density.
The distribution of pixel HU at different axial slices for sample patient images are shown in Fig. 5. Typical Radio-densities in HU of various materials in CT scan images of the lung are given in Table 3.
To exclude those tissues, some segmentation methods extract the mask of lung and leave all other tissues in the detection stage. A common approach used by researchers are thresholding (Alakwaa et al. 2017), clustering (Rao et al. 2016), watershed (Ronneberger et al. 2015) and k-means (Gurcan et al. 2002). For each 2D slice, we used thresholding to filter 2D image with Gaussian filter and then normalize the pixel value to [0, 1] using − 600 as a threshold. An original 2D CT slice of a sample patient and the resulting 2D slice of the lung segmentation mask created by thresholding is shown in Fig. 4.
To prepare the data for the network, we shift the image from HU to UINT8. The raw data are initially clipped within [− 1024, 3071], and linearly shift to [0, 255] (Fig. 5).
3.2.2 Nodule detection
Following Rao et al. (2016), we train a modified version of U-Net (we modify some parameters of U-Net, such as input size, kernel size, model depth.) on LUNA 16 data to segment suspicious nodules. U-Net is a 2D model that is widely applied to various Bio-medical image segmentation tasks, such as brain tumor segmentation, ultra-sound nerve segmentation, and retina blood vessel segmentation. For more understanding about U-Net, we refer the reader to read (Ronneberger et al. 2015). The parameters of the modified U-Net are given in Table 2.
During training, the modified U-Net takes as an input 256 × 256 2D CT slices, and their corresponding labels are provided by masking 256 × 256, where nodule pixels are 1 and the rests are 0.
The output of the model is an image having the same size with an input. Each pixel of the output has a value between 0 and 1, showing the probability the pixel belongs to a nodule. This is utilized by taking the slice belongs to label 1 of the softmax of the final U-Net layer.
Finally, the trained U-Net is then used to segment the KDSB CT scan slices. These candidates have variable size (small, medium and large) and shape (circular, elliptical and others), where we categorized them into training set, validation set, and test set to train the proposed mp-CNN (Table 3).
4 Proposed multi-path CNN method
4.1 CNNs
The core building block used to design a CNN architecture is the convolution layer. Many layers can be heaped on top of each other creating a hierarchy of features. Each layer can be understood as extracting features from its previous layer into the structure to which it is associated. A convolution layer takes as input a heap of input patches and gives as output some number of planes called feature maps. Each feature map \( f_{j} \) is associated with one weight. Basically computing a feature map in a convolution layer has three steps (Fig. 6).
First, given an \( i \)-th input channel \( x_{i} \) with \( w_{j,i} \) a sub-weight of that channel and biases term \( b_{j} \), the feature map \( f_{j} \) using convolution is computed as
where \( * \) is a convolution operation. The key to the achievement of CNN is their capability to learn the weight and biases of separate features, giving boost to data-driven customized task-specific compact features. These parameters are optimized through stochastic gradient descent on an alternate loss function related to the misdetection, with gradient calculated through backpropagation algorithm (Rumelhart et al. 1988). Second, to get features that are nonlinear transformation of the input, an element wise operation is applied to the result of the kernel convolution. There are various choices of such function. Recently, ReLu defined as
were found to attain better results than the more conventional sigmoid, or hyperbolic tangent functions, and also help in speeding up the training process (Ramachandran et al. 2017; Jarrett 2009). Sometimes, imposing a constant zero can harm the gradient flowing and subsequent adjustment of the weights. Due to this some researchers used leaky rectifier linear unit (lrelu) (Maas et al. 2013)
that presents a small slope on the negative part of the function, where \( \alpha \) is the leakiness parameter. However, during our experimentation we have not obtained substantial change by the use of lrelu, and thus we used Eq. (2). Third, to shrink the size of feature maps, pooling is applied on each feature map. Max-pooling and average pooling are commonly used in medical image detection.
From the inference of neural networks, feature maps are corresponding to a layer of hidden neurons. Each coordinate of the feature maps is corresponding to an individual units or neurons, for which the size of its receptive field corresponding to the filter size. The filter value indicates the weight of the connection between the layer’s neurons in the previous layer.
Finally, to achieve an indicator of the detection labels, one can associate the last convolution hidden layer to a convolutional output layer followed by a nonlinearity activation function without pooling operation. In most CNN detection models, the output layer is normally fully connected, however one can redesign the CNN architecture subject to a specific task. In this paper, the last output layer is connected to a convolutional layer without fully connected. With this adapted CNN architecture, contested to the traditional CNN architectures that employs fully connected layers, we observed that the computational time is improved. We also made several modifications, and hence observed an improvement over the conventional one. Many weights are used; these weights acts as the final detector of nodules from one of the detection labels. We use the softmax which normalize into two distributions over the labels. Particularly, assume \( x \) be the vector of values at a spatial position, it calculates softmax \( s(x) \)
where z is normalized constant and
Assuming \( Y \) as a detection label over the input patch \( X \), we can thus interpret each spatial position of a convolutional output layer as bringing a model for the likelihood distribution \( P(Y_{i,j} |X) \) where \( Y_{i,j} \) is the label at coordinate \( Y_{i,j} \). We obtain the probability of all labels easily by accepting the product of each conditional probability \( P(Y|X) = \prod\nolimits_{i,j} {P(Y_{i,j} |X)} \). Our method hence performs binary class labeling by assigning to each pixel the label with the largest probability.
4.2 Multi-path CNN architecture
So far, our explanation of CNN’s suggests a simple architecture belonging to a single heap of several convolutional layers. In the field of computer vision, this architecture is the most commonly implemented architecture. However, one could design other architecture that might be suitable for the problem at hand.
As we have mentioned earlier, some of the key challenges of lung cancer detection using CNN are the variability among nodules size, shape, and their contextual characteristics. Due to these challenges, the existing CNN based lung cancer detection models face the problem of accuracy. One reason for the aforementioned problems is that the receptive field size influence while modeling distant dependencies. Most CNN based lung cancer detection relies on fixed receptive field size (example, the use of 3 × 3 or 5 × 5 size throughout the architecture) which affect the contextual and visual information’s of neighborhood pixels during feature extraction.
In this paper, to circumvent the problem, we design a CNN having multi-path convolutional layers, i.e., the path considering smaller, medium, and larger receptive field sizes. We call these paths first, second, and third path, respectively.
The receptive field size of the first path is 3 × 3, the second path is 5 × 5, and the third paths is 7 × 7. Here one can ask as if the receptive field size 7 × 7 larger or 3 × 3 is appropriate enough? The answer is subjective; it depends on a task. We just focus on these receptive field sizes to study the task at hand. Moreover, the design of our architecture is supported by the concatenation of different feature maps from the last convolutional layers of the three paths which help the model to boost the prediction.
The motivation for this architecture design is that we would like the prediction of the label of a pixel to be positively influenced by:
-
the visual details of the region around neighborhood pixels and
-
contextual information.
Also, the concatenation layer helps to determine important features from the three pathway which helps in modeling long and short-term dependencies, i.e., features in the same areas are modeled in a better way. We observed that this architecture overcome the effects of receptive field size and provided better accuracy contested to the traditional CNN’s model which employ fixed receptive size and one way convolutional layers. The full architecture is illustrated in Fig. 7 and its parameters are given in Table 4.
Multi-pathway CNN architecture. The figure demonstrates the input patch move through three paths of convolutional operations. The feature maps in the first, second and third paths are shown in blue, red, and green colors, respectively. The convolutional layers used to output these feature maps are indicated in Table 4 (Color figure online)
We refer to this architecture as multi-path CNN (mp-CNN). We describe these three paths of mp-CNN architecture that we used in this work one by one.
4.2.1 First path
This CNN path is the path with smaller 3 × 3 receptive field size intended to model short-range dependencies of a neighboring pixel. It has four convolutional layers where the first three layers are followed by 2 × 2 max pooling and the fourth layer is followed by 4 × 4 max pooling. The 4 × 4 max pooling is applied because we want the same spatial size of feature maps with other paths. It helps us to easily concatenate feature maps. The first convolutional layer takes a preprocessed 2D slices of lung images of size 128 × 128 × 1 as input and filtered with 64 filters of size 3 × 3. The second layer takes the output of the first layer (after max-pooling) and performs convolution, where 64 kernels of size 3 × 3 are applied to feature maps of size 64 × 125 × 125. Similarly, after max pooling, the third layer takes the output of the second layer as an input and perform convolution with 64 filters of size 3 × 3 on feature maps of size 64 × 122 × 122. Finally, the fourth layer of this path takes the output of the third layer of size 64 × 119 × 119 as input and perform convolution with 64 filters of size 3 × 3. Clearly, this layer has 64 × 114 × 114 output size, after 4 × 4 max pooling. The output of the third layer is then concatenated together with the output of the last layer of the remaining two paths.
4.2.2 Second path
This is a path with medium 5 × 5 receptive field size intended to model average short and long-range dependencies of neighbor pixels. The medium sized and various shaped of lung nodules are better modeled through this path, where the larger and smaller nodules are better modeled through larger receptive and smaller receptive field respectively, and also concatenation from the three paths boost performance. The path has three convolutional layers, each of them are followed by 2 × 2 max pooling. Similar to the first path, the first layer of this path takes a preprocessed 2D slices of lung cancer as an input and 64 filters of size 5 × 5 are applied to it. After max poling is applied to the output of the previous layer, the output becomes the input to the second convolutional layer. It takes 64 × 123 × 123 spatial size as input, 64 filters of size 5 × 5 are applied to this input which then outputs feature maps of size 64 × 119 × 119 (i.e., 64 feature maps each 119 × 119 spatial size), after max pooling. Then, the next layer (i.e., layer three) takes the outputs of the previous layer and convolution is applied to it with 64 filters of size 5 × 5 again.
4.2.3 Third path
This path is aimed to capture long-range dependencies. It has two convolutional layers followed by max pooling of size 2 × 2. The first layer takes 128 × 128 × 1 as inputs, convolution is applied on this input with 64 filters of size 7 × 7 and outputs 64 × 122 × 122 feature maps. After max pooling, it becomes the input to the second layer. The second layer also applies the same operation with 64 filters of size 7 × 7 and output 64 × 115 × 115. After 2 × 2 max pooling, its output is concatenated together with the output of the last convolutional layer of the other two paths.
4.2.4 Concatenation
To average the effect of receptive field size, we concatenate the output of the last convolution layer of each path, and this concatenation is convolved with 192 filters of size 114 × 114 and then followed by a soft-max function to predict the input.
Generally, the use of multi-path model with concatenation both exploit the efficiency of CNN’s while also more exactly model the dependencies between neighbor pixels in the detection task.
Such joint detection model is more computationally expensive than a feedforward passes via CNN. This is a very crucial step when we should take into consideration if automatic lung cancer detection is to be used in a day to day work.
Here we explain the CNN model that both feat the efficiency of CNN’s, while model the distant dependencies. Since we would like the final prediction to be prejudiced by the model’s views about the neighborhoods labels, we put forward to feed the output probabilities of a third and the second CNN as additional inputs to the layer of the first CNN. We do this through a concatenation of convolutional layers. In this case, we concatenate the last convolutional output layers of each path of CNN.
4.3 Training
By describing the output of the CNN as a model for the distribution over detection labels, a common training rule is to maximize the probability of all labels in training set or, to minimize negative log-probability
for each labeled lung slice.
To perform this, we use stochastic gradient method by repeatedly choosing labels \( Y_{i,j} \) at a random subset of patches within each lung, calculating the mean negative log-probabilities for this mini batch of patches and doing a gradient descent step on the CNNs parameters (i.e., the weight at all layers). We perform updates only on small subset of patches. This allows us to avert having to process the whole slice of the lung for each update, at the same time providing decent updates for learning. Particularly, we perform this method by forming a dataset of mini batches of smaller slices of lungs image patches, paired with the analogous detection label as the target.
Since momentum tactic has been fruitful in the past (Krizhevsky et al. 2012), we implement it to optimize our model. We used this strategy as
where \( W_{i} \) represents the CNNs parameters at iteration \( i \), \( \nabla{W_{i}} \) is the gradient of the loss function at \( W_{i} \), \( v_{i} \) is the integrated velocity where the initial value is set to zero, \( \gamma \) is the learning rate, and \( \eta \) is the momentum coefficient. We set the initial momentum \( \eta = 0.5 \) and the final momentum \( \eta = 0.9 \). Also, learning rate \( \gamma = 0.005 \) is decayed exponentially with decay factor 0.1.
5 Experimental result and discussion
5.1 Experimental setup
We implement an experiment on one of the deep learning library- tensor flow (Martin 2016) with Kera backend. It supports GPUs which can highly accelerate the computation of deep learning algorithm. Hyper-parameters of our model such as filter size, max pooling size, architecture depth, and others are given in Table 4. These chosen hyper-parameters were the one for which our models performed best on the validation set. For both convolutional and max-pooling layers, we used a stride 1. This helps us to keep the per-pixel accuracy. From our experiment, we observed that adding additional convolutional and max-pooling layers on the third path does not improve the results, also we found adding additional features does not give any significant improvement. That is why we limit the layer depth of our model. Weights are randomly initialized from the uniform distribution \( U( - 0.005, - 0.005) \) and except for the softmax layer for which we initialized them to the log of the label, biases are set to zero.
5.2 Evaluation metrics
To measure how our model well perform, we compute the commonly used image detection performance measures accuracy, \( A \), specificity, \( S \), recall, \( R \):
where \( tp \) is the number of true positives, \( fp \) is the number of false positives, \( fn \) is the number of false negatives, and \( tn \) is the number of true negative. One can multiply these equations by 100 to give the results in percentage.
5.3 The preprocess stage
Initially, we have trained our model with the raw KDSB dataset without lung segmentation and nodule detection. In addition to the shortage of images that KDSB comprises, unnecessary substances that exist in the CT scan image of the lung were not removed, and due to these we obtained bad results. Then we removed tissues like bone, air, water, and trained the mp-CNN, still, the results were not satisfactory. Hence, to improve our model performance, we performed lung segmentation and nodules detection on the raw images. Thresholding was applied to segment the lung, and then, U-Net was used to detect suspicious nodules. The trained U-Net model was used to detect the region where the suspicious nodules (cancer or non-cancer nodules) would be available (KDSB dataset was used). Experimental result shows, the use of Thresholding and U-Net before directly employing multi-path convolutional neural network improved the mp-CNN performance. Table 5 and Fig. 8 shows the results of our model accuracy with and without preprocessing stage. As shown in the table, the accuracy of mp-CNN improved because of the preprocessing stage applied. Also, from Fig. 8 one can observe that mp-CNN model accuracy is increasing over the number of training steps when preprocessing stage is performed. mp-CNN trained with the preprocessing stage has on average 3.6% accuracy than mp-CNN that was not trained with the aforementioned preprocessing. From the experiments, we noticed that the joint segmentation and detection preprocess exploited were improved the proposed mp-CNN performance.
5.4 The multi-path architecture
As we have mentioned earlier, unlike the traditional CNN, the mp-CNN has three pathways. These paths are designed to better approximate local and global dependencies of the neighboring pixels. The first and the second path more focused on the details (local dependency) and the third path focused on the contextual information (global dependency).
Our focus on both contexts and details lead the model for better performance. To better interpret how joint training of these paths helps the performance, we address results on each pathway and also results on averaging the output of each pathway when trained independently.
Since we also deal with the labeled dataset class imbalance, we retrain the model with an approach described in Sect. 3.1. To see the consequence of the two-phase of training, we address the results of both, i.e., the results of the first training phase and the results of the retraining phase. We refer our CNN model consisting of only first path, second path, and third path as f-CNN, s-CNN, and t-CNN, respectively. Also, the CNN model averaging the output of the three paths as a-CNN and the overall multi-path CNN model as mp-CNN. The second training phase is noted by superscript ‘s’.
Table 6 shows the quantitative results of these alterations. The table comprises results for the mp-CNN and f-CNN with single and retraining phase, and s-CNN, t-CNN and a-CNN with only retraining phase. As one can observe from the table, the first path with one training phase ranked as last, however, using a retraining phase provided a substantial improvement to that model. In addition, if we observe the performance of mp-CNN without retraining phase, it provided less performance than f-CNNs, s-CNNs, and a-CNNs, but after the retraining phase, mp-CNN ranked first.
This reveals that:
-
collaborative training of the first, second, and third paths delivers better performance contested to when each pathway is trained independently and the output is averaged.
-
the retraining phase plays a substantial role to improve the mp-CNN performance.
Indeed, a-CNNs performs lower than f-CNNs because we believe t-CNN performs badly. The mp-CNNs is the best performer model.
Table 7 reveals the performance results of our model versus the currently published state of the art lung cancer detection methods. The table reveals that mp-CNNs outperformed all of them. It gains 0.016 recall, 0.010 specificity, and 0.014 accuracy values over (Kingsley et al. 2017), and provided greater recall, specificity, and accuracy values over (Alakwaa et al. 2017; Rao et al. 2016; Huang et al. 2017). Even the f-CNNs determined better recall, accuracy, and specificity results than (Kingsley et al. 2017; Alakwaa et al. 2017; Rao et al. 2016; Huang et al. 2017). To make more specific our model has provided competitive results compared to the winner of the Kaggle Data Science Bowl 2017 challenge.
5.5 Shape and size effect
We selected 50 large cancer nodules and 50 small size cancer nodules having various shapes from KDSB 2017 challenge and evaluated our model performance. Table 8 shows the obtained results. As one can observe from the table, of 50 larger cancer nodules selected, 98% were correctly predicted with our mp-CNNs model, and of 50 smaller cancer nodules selected, 94% were correctly predicted. Moreover, our model better performs on larger nodules than the smaller nodules. Compared to some other lung cancer detection methods (Kingsley et al. 2017; Alakwaa et al. 2017; Rao et al. 2016; Huang et al. 2017), our mp-CNNs has better performance results. In general, we have shown that the proposed mp-CNN for lung cancer detection better address the problem of variability among lung nodules.
6 Conclusion
In this paper, we introduced an automatic lung cancer detection method using deep convolutional neural networks. We considered various architecture and analyzed their effect on the detection performance.
Experimental result conducted on KDSB 2017 approves that with our best model we managed to improve on the currently published state-of-the-art method on accuracy, sensitivity and specificity.
The greater performance is obtained with the help of a novel multi-pathway architecture, which can model various size and shape of the lung nodules, because short and long-range dependencies (details and context) are modeled in an appropriate way. Training procedure has two phases which helped us to train CNN’s effectively and efficiently when the distribution of labels class is unbalanced. Because of the nature of CNN models, by GPU machine, the resulting detection is fast.
In general, compared to other lung cancer detection methods using CNN models, our model is flexible, can better handle the variation of size and shape of nodules and balance the labels class imbalance.
References
Alakwaa, W., Nassef, M., & Badr, A. (2017). Deep convolutional neural networks for lung cancer detection. IJACSA, 8(8), 403–417.
Alvarez, J. M., Gevers, T., LeCun, Y., & Lopez, A. M. (2012). Road scene segmentation from a single image. In Proceedings of the 12th European conference on computer vision (pp. 376–389).
Clark, M., Hall, L., Goldgof, D., Velthuizen, R. P., Murtagh, F., & Silbiger, M. L. (1998). Automatic tumor segmentation using knowledge-based clustering. IEEE Transactions on Medical Imaging, 17, 187–201.
Gurcan, M. N., Sahiner, B., Petrick, N., Chan, H. P., Kazerooni, E. A., Cascade, P. N., et al. (2002). Lung nodule detection on thoracic computed tomography images: Preliminary evaluation of a computeraided diagnosis system. Medical Physics, 29(11), 25522558.
Havaei, M., Davy, A., Warde-Farley, D., Biard, A., Courville, A., Bengio, Y., et al. (2017). Brain tumor segmentation with deep neural networks. Medical Image Analysis, 35, 18–31.
Huang, X., Shan, J., & Vaidya, V. (2017). Lung nodules detection in CT using 3D Convolutional neural networks. In International Symposium on IEEE (pp. 379–383).
Jarrett, K., et al., (2009). What is the best multi-stage architecture for object recognition? In Proceedings 12th International Conference on IEEE Computer Vision (pp. 2146–2153).
Kaggle Data Science Bowl (KDSB). (2017). https://www.kaggle.com/c/data-science-bowl-2017/data. Accessed 25 Mar 2018.
Kingsley, K., Mathieu, R., Gaurav, M., Huiling, C., Jie, L., Babar, N., et al. (2017). Deep learning for lung cancer detection: Tackling the Kaggle data science bowl 2017 challenge, arXiv preprint arXiv:1705.09435v1.
Krizhevsky, A., Sutskever, I., & Hinton, G. (2012). Image Net classification with deep convolutional neural networks. In NIPS.
Lepor, H. (2000). Prostatic diseases. Philadelphia: W.B Saunders Company.
Lin, D. T., & Yan, C. R. (2002). Lung nodules identification rules extraction with neural fuzzy network. International Conference on in Neural Information Processing, 4, 20492053.
Liu, W., Anguelov, D., Erhan, D., Szegedy, C. Reed, S., Fu, C.-Y., et al. (2016). SSD: single shot multi box detector. In European Conference on Computer Vision (pp. 21–37).
Liu, Y., Gadepalli, K., Norouzi, M., Dahl, G. E., Kohlberger, T., Boyko, A., et al. (2017). Detecting cancer metastases on giga pixel pathology images, arXiv preprint arXiv:1703.02442.
Lung Nodule Analysis (LUNA). (2016). https://luna16.grand-challenge.org/. Accessed 2 Mar 2018.
Maas, A. L., Hannun, A. Y., & Ng, A. Y. (2013). Rectifier non linearity improve neural network acoustic models. In Proceedings of ICML (Vol. 30).
Martin, A. et al. (2016). TensorFlow: Large-scale machine learning on heterogeneous distributed systems, arXiv preprint arXiv:1603.04467.
Muhammad, R., Saeeda, N., & Ahmad, Z. (2017). Deep learning for medical image processing: Overview, challenges and future, arXiv preprint arXiv:1704.06825.
Patz, E. F., Jr., Pinsky, P., Gatsonis, C., et al. (2014). Over diagnosis in low-dose computed tomography screening for lung cancer. Internal Medicine, 174(2), 269–274.
Pereira, S., Pinto, A., Alves, V., & Silva, C. A. (2016). Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Transactions on Medical Imaging, 35(5), 1240–1251.
Ramachandran, P., Zoph, B., & Le, Q. V. (2017). Searching for activation functions, arXiv preprint arXiv:1710.05941v2 [cs.NE].
Rao, P., Pereira, N. A., & Srinivasan, R. (2016). Convolutional neural networks for lung cancer screening in computed tomography (CT) scans. In International conference on contemporary computing and informatics (pp. 489–493).
Rebecca, L. S., Kimberly, D. M., & Ahmedin, J. (2018). Cancer statistics, 2018. CA: A Cancer Journal for Clinicians. https://doi.org/10.3322/caac.21442.
Redmon, J., & Farhadi, A. (2016). Yolo: Better, faster, stronger, arXiv preprint arXiv:1612.08242.
Ren, S., He, K., Girshick, R., & Sun, J. (2015). Faster R-CNN: Towards real-time object detection with region proposal networks. In Advances in Neural Information Processing Systems (pp. 91–99).
Ronneberger, O., Fischer, P., & Brox, T. (2015). U-Net: Convolutional networks for biomedical image segmentation, arXiv preprint arXiv:1505.04597.
Rumelhart, D. E., Hinton, G. E., & Williams, R. J. (1988). Learning representations by back-propagating errors. Nature, 323(6088), 533–536.
The National Lung Screening Trial Research Team. (2011). Reduced lung-cancer mortality with low-dose computed tomographic screening. New England Journal of Medicine, 365(5), 395–409.
WHO. (2018). http://www.who.int/news-room/fact-sheets/detail/cancer. Accessed 26 Oct 2018.
Acknowledgements
This work is partially funded by the MOE-Microsoft Key Laboratory of Natural Language Processing and Speech, Harbin Institute of Technology, the Major State Basic Research Development Program of China (973 Program 2015CB351804), the National Natural Science Foundation of China under Grant Nos. 61572155, 61672188 and 61272386 and Bule Hora University, Ethiopia. We would also like to acknowledge NVIDIA Corporation who kindly provided two sets of GPU. We would like to acknowledge the editors and the anonymous reviewers whose important comments and suggestions led to greatly improved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sori, W.J., Feng, J. & Liu, S. Multi-path convolutional neural network for lung cancer detection. Multidim Syst Sign Process 30, 1749–1768 (2019). https://doi.org/10.1007/s11045-018-0626-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11045-018-0626-9