Abstract
Colorectal cancer refers to cancer of the colon or rectum; and has high incidence rates worldwide. Colorectal cancer most often occurs in the form of adenocarcinoma, which is known to arise from adenoma, a precancerous lesion. In general, colorectal tissue collected through a colonoscopy is prepared on glass slides and diagnosed by a pathologist through a microscopic examination. In the pathological diagnosis, an adenoma is relatively easy to diagnose because the proliferation of epithelial cells is simple and exhibits distinct changes compared to normal tissue. Conversely, in the case of adenocarcinoma, the degree of fusion and proliferation of epithelial cells is complex and shows continuity. Thus, it takes a considerable amount of time to diagnose adenocarcinoma and classify the degree of differentiation, and discordant diagnoses may arise between the examining pathologists. To address these difficulties, this study performed pathological examinations of colorectal tissues based on deep learning. The approach was tested experimentally with images obtained via colonoscopic biopsy from Gyeongsang National University Changwon Hospital from March 1, 2016, to April 30, 2019. Accordingly, this study demonstrates that deep learning can perform a detailed classification of colorectal tissues, including colorectal cancer. To the best of our knowledge, there is no previous study which has conducted a similarly detailed feasibility analysis of a deep learning-based colorectal cancer classification solution.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Deep learning has gained much attention from various research communities as it has been able to solve many complex tasks with significantly higher accuracy than conventional techniques [11]. In particular, deep learning has been extensively employed in various fields of healthcare for tasks such as classification and localization of diseases from various modalities, segmentation of anatomical structure, survival analysis, and drug discovery. Comprehensive surveys on medical deep learning are provided in [5, 10, 12, 13]. In some cases deep learning-based solutions have outperformed the expert human level by reducing the errors caused by inter and intra-human variations. Subsequently, this resulted into increased efficiency and more objective results. As such, deep learning has been successfully applied in computer vision in recent years and in various applications across several areas. For example, convolutional neural networks (CNNs) have shown high recognition rates of human handwriting [16]. Moreover, numerous studies have demonstrated the applicability of deep learning in real-life applications, and various studies on medical image analysis through deep learning are also underway [12]. For instance, deep learning has been applied to breast cancer [21] and glaucoma diagnoses [9], resulting in faster and more accurate examination than that of existing image processing techniques. Accordingly, this study performed pathological examinations of colorectal tissues based on deep learning.
Colorectal cancer has the world’s third-highest cancer incidence rate after lung cancer and breast cancer, and is the second leading cause of cancer deaths [2]. Typically, the diagnosis of colorectal cancer is mainly done by the biopsy of colorectal tissue through colonoscopy, which is processed, dyed, and prepared on glass slides. Subsequently, a pathologist diagnoses the slides through a microscopic examination. The size, shape, and location of the colorectal tissue observed by pathologists vary widely. As shown in Fig. 1, for normal colonic mucosa, the number and arrangement of epithelial cells remain relatively constant, and as it progresses to adenoma and adenocarcinoma, the epithelial cells overproliferate and form complex structures. This makes the pathologic examination lengthy and labor-intensive, and it may sometimes be difficult to obtain consistent diagnoses owing to conflicting opinions between pathologists. Therefore, automated analysis by computers that are consistent and objective may play a crucial role in improving diagnostic accuracy by helping pathologists make microscopic examinations.
Automated analysis solutions for colorectal tissue based on traditional image processing techniques [3, 20] have several limitations. such as an ingerent vulnerability to image noise and relatively lower capability of generalization. To address these issues, researchers have conducted numerous studies on colorectal tissue examination using deep learning. Using a VGG model [15], Francesco Ponzio et al. [14] performed transfer learning [19] to classify samples into normal tissues, adenomas, and adenocarcinomas, and the authors obtained a certain level of accuracy. Classifying samples into three classes (normal tissue, low-grade adenocarcinoma, and high-grade adenocarcinoma), Ruqaya Awan et al. [1] used a CNN model to detect and classify the gland outlines observed in colorectal tissues. Accordingly, this study hypothesized that deep learning could classify various colorectal tissues histologically. For this purpose, ResNet [7], DenseNet [8], and Inception V3 [18], models with excellent feature extraction performance, were trained to classify colorectal tissues. According to the training results, all models exhibited a remarkable performance of more than 0.9 mAP(mean average precision). The objective of this paper is to demonstrate the feasibility of deep learning based solution for classifying five different cases of colorectal cancers (i.e., Normal, Adenoma, Adenocarcinoma-well, Adenocarcinoma-moderate, and Adenocarcinoma-poor), which, to the best of our knowledge, has not yet been recorded. In particular, none of the existing work has dealt with three detailed classes of adenocarcinoma.
The paper is structured as follows. Section 2 describes related works, whereas Section 3 describes the extraction method of the dataset and the CNN used for training. Section 4 describes the experimental process and results. Finally, Section 5 concludes the study and discusses future works.
2 Related works
Simon Graham et al. [6] segmented the gland epithelial cells for all slide images of a colorectal cancer dataset. By so doing, the authors helped to reduce the difficulty of manual segmentation and uncertain decision-making in pathology. To perform this study, the researchers proposed a complete CNN that re-introduces the original images at numerous points in the network to compensate for information loss due to max pooling.
Francesco Ponzio et al. classified colorectal tissues into the three types, namely normal, adenoma, and adenocarcinoma. For the experimental data, they extracted 109 areas using 27 original slides and generated 13,500 image patches from the areas. The authors then used a VGG 16 model where the colorectal tissues were not relatively deep, demonstrating the feasibility of the classification with a certain level of accuracy.
Ruqaya Awan et al. stated that the extent of gland formation determines the grade of adenocarcinoma. They measured the extent of gland formation using the ”best alignment metric” (BAM), a new measurement method, and classified colorectal tissues into normal, low-grade cancer (well- differentiated and moderately differentiated), and high-grade cancer (poorly differentiated and undifferentiated). The study demonstrated that the grade of adenocarcinoma could be classified by detecting the outlines of glands.
Nassima Dif and Zakaria Elberichi[4] proposed a dynamic ensemble deep learning method to address the limitation of deep learning methods in histopathological image analysis owing to the restricted number of medical images available for training. They applied the particle swarm optimization algorithm to select component models which were then ensembled by averaging or voting methods.
The present study performed a detailed adenocarcinoma classification and categorized colorectal tissues into a normal, adenoma, and adenocarcinoma (well differentiated, moderately differentiated, poorly differentiated). This study presents a more precise classification method than the colorectal tissue classifications methods of previous studies using deep learning. Furthermore, as this study used 693 original data slides, a large number of colorectal tissues were studied compared to previous studies.
3 Data set and method
3.1 Datasets
The colorectal tissue images were selectively obtained by scanning glass slides prepared from 850 colorectal tissues collected via colonoscopy from Gyeongsang National University Changwon Hospital from March 1, 2016, to April 30, 2019. This study was approved by the Institutional Review Board of Gyeongsang National University Changwon Hospital with a waiver for informed consent (2019-05-008) Moreover, through the examination of an experienced pathologist, the colorectal tissues were classified into normal, adenoma, and adenocarcinoma, and, in cases of adenocarcinoma, the tissues were classified into three differentiation types of well differentiated, moderately differentiated, and poorly differentiated, based on which the data were labeled. The degree of differentiation was determined by how much the shape of the gland was maintained: well differentiated if maintained at more than 95%, moderately differentiated at 50% to 95%, and poorly differentiated at less than 50%. The training and test data patches for each class were composed, as shown in Table 1.
To facilitate an efficient learning process, the training data slides were processed and composed in two steps. In the first step, only the parts in the original slides (Fig. 2a) annotated according to the classification of the pathologist were manually extracted into the areas shown in Fig. 2b. In the second step, the image patches were extracted from the areas obtained in the first step. Moreover, the processed areas were treated in the form shown in Fig. 3a, using the algorithm presented in Fig. 4, in which it is assumed that patch images are square rectangles. That is to say, the parts overlapping (IOU) with the subsequently processed data are set to 0.3 and moved from the upper-left to the right, similar to Fig. 3a. The training data is cropped to a size of 400 x 400 (lines 10–13).
Once the data in one row are completely extracted, it is moved to the next row (line 9, 14). Here, the portions where the data of the upper and lower rows overlap are also set to 0.3, and as in the previous row, the data are moved to the right and extracted. However, when creating the training data patches using the sliding window technique, the data extraction method was very formal, leading to limitations in creating data of various characteristics. Accordingly, additional training data, 400 x 400 in size, were randomly extracted from the area data without using the sliding window technique, thus expanding the amount of data. Moreover, the data containing numerous features were extracted in the form shown in Fig. 3b. Among the removed data patches, those that did not contain at least 50% of glands were excluded from the training data. The test data were constructed in a manner similar to that of the training data and processed using the area data excluded from the training data.
3.2 Method
This study classified the colorectal tissues using the ResNet, DenseNet, and Inception V3 CNN models, which have demonstrated excellent performance in data feature extraction. The core module of ResNet is the structured residual block shown in Fig. 5a. The flow between the weight layers of the existing CNN is a structure that passes the weight layers and extracts and outputs features for a given input value x.
However, the residual block uses a skip connection that directly connects the input x to the weight layers. Whereas the existing CNN finds output values from the input value, here, the learning is performed to minimize the value of F(x) using a skip connection. This results in fast learning speeds and avoids increasing the number of operations, since the skip connection only adds a simple addition operation. Accordingly, this study used a model constituting a 50-layer ResNet.
DenseNet continuously connects each layer with the input of the next layer through the feature map, as shown in Fig. 5b. Though ResNet uses a similar technique, the greatest difference is that ResNet has a structure that adds feature maps, whereas DenseNet has a stacked structure. This structure can stack information of the preceding layer and efficiently transfer it to the subsequent layer, which improves the vanishing gradient and strengthens feature propagation and feature reuse without relearning the same features, thereby reducing the number of parameters. This study used a model constituting a 121-layer DenseNet.
Inception V1[17] uses the Inception module to solve the problems of gradient vanish and overfitting, in which, as the network depth increases, the parameters substantially increase, and the computational quantity exponentially increases. As shown in Fig. 5c, Inception V2 model was presented that can further reduce the computational cost by combining three modules: a module replacing the 5 × 5 convolution with two 3 × 3 convolution operations, a module replacing the 3 × 3 convolution with a 1 × 3 convolution and 3 × 1 convolution, and a module widening the Inception module to solve the representational bottleneck problem. While Inception V3, which is used in this study, has the same structure as Inception V2, the Inception module in Inception V3 replaces the 7 × 7 convolution with three 3 × 3 convolutions, changes the optimizer to RMSProp, and uses batch normalization in the last completely connected layer and label smoothing.
4 Experiments
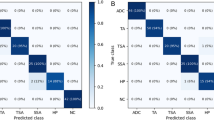
Tensorflow and Python 3.6 were used to implement each CNN model used in this study, and they were trained for 15 hours using a Geforce RTX 2080 GPU. The batch size of ResNet, DenseNet, and Inception V3 was set to 32, and learning was performed for 200 epochs. The loss value quickly decreased at the start of learning and converged as the learning progressed. The learning rate was initially set to 1e-3, and was subsequently gradually decreassd to 1e-4, 1e-5, 1e-6, 1e-7 after 80, 120, 160, and 180 epochs, respectively. For optimization, the Adam method was used as the optimizers for ResNet and DenseNet, while RMSProp was used for Inception V3. The number of output layers of the three CNN models was set to 5, and learning was performed. Figure 6 shows that the losses of the three CNN models converged, indicating that learning was successfully performed. Figure 7 shows the AUROC(Area Under Receiver Operating Characteristic) curves of the three models used in the experiment. According to the tests, all three models showed performances of more than 0.9 AUC for each type of colorectal tissue, as shown in Fig. 7. However, as shown in the confusion matrix in Fig. 8, the colorectal tissue glands are continuous and irregular. Therefore, in each trained CNN model during testing, some parts are slightly indistinguishable from the adjacent colorectal tissues for each type. The performance metrics were compared with those of Table 2. Moreover, Table 3 shows the overall performance of the three models. The ResNet model exhibited the best performance of the three. However, as shown in Table 4, for well differentiated, moderately differentiated, and adenoma, DenseNet and Inception V3 partially exhibited excellent performance. As precision and recall have a trade-off relationship, the DenseNet precision of the well differentiated parts was relatively poor compared to that of the other models. However, the ResNet model exhibited excellent precision and recall performance for poorly differentiated parts.
Figure 9 shows the judgment positions for classifying the colorectal tissues using the ResNet-trained model through the CAM(Class Activation Map) technique[22]. The five images in the first row of each colorectal cancer type are data patches of the test set, and the five data patches in the second row are parts showing the judgment positions through the trained model. In the data patches with differing brightness in the second row, brighter portions had a greater impact on the trained model decision.
The five data patches in the last row of Fig. 9 are combinations of the test data patches in the first row and images using the CAM technique in the second row. As such, they show combinations of the colorectal tissue data and CAM technique patches. The model is trained from five images using the CAM technique for each type and detects the gland shape and size of the cell membrane and nucleus, which are parts that greatly impact the model’s decision. Pathologists also diagnose colorectal tissues using the shape of the epithelial cell’s nucleus and the degree of gland fusion. Therefore, the classification of colorectal tissues by a trained model using the CAM technique is consistent with the pathologists’ criteria. This demonstrates that the trained models in this study accurately identify the features of colorectal tissues.
In some test data patches, however, some parts greatly affected the model’s decision which differed from the pathologists’ criteria (see Fig. 10). The arrows in Fig. 10 indicate the parts with the greatest influence on colorectal tissue classification in the trained model, showing that the model only looks at certain parts. While these parts are within the scope of the examination criteria, there are limitations in looking at specific parts rather than the entire area.
5 Conclusion and future planning
This study presented the feasibility of applying deep learning in the classification of colorectal tissues. As such, this study approached colorectal tissue classification as an image classification problem, which is a computer vision process. Moreover, this study applied deep learning to image analysis. The training was performed using three CNN models to verify the performance, and they all exhibited excellent performance. According to the experimental results, the judgment positions of the model trained using the CAM technique were identical to the pathologists’ criteria for most of the test data patches.
This was a retrospective study of a single institution using data from colorectal tissue slides of Gyeongsang National University Changwon Hospital. There are limitations in the slide photographs acquired from an external institution using processing techniques and machines with different models. To generalize this study, contivuing research should focus on processing additional training data using slides acquired through various methods and processing techniques. Moreover, further studies shall be conducted to make judgments for data considering the limitations.
References
Awan R, Sirinukunwattana K, Epstein D, Jefferyes S, Qidwai U, Aftab Z, Mujeeb I, Snead D, Rajpoot N (2017) Glandular morphometrics for objective grading of colorectal adenocarcinoma histology images. Scientific Reports 7(1):1–12
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a Cancer Journal for Clinicians 68 (6):394–424
Canny J (1986) A computational approach to edge detection. IEEE Transactions on Pattern Analysis and Machine Intelligence 8(6):679–698
Dif N, Elberrichi Z (2020) A new deep learning model selection method for colorectal cancer classification. International Journal of Swarm Intelligence Research (IJSIR) 11(3):72–88
Egger J, Gsxaner C, Pepe A, Li J (2020) Medical deep learning–a systematic meta-review. arXiv:201014881
Graham S, Chen H, Gamper J, Dou Q, Heng PA, Snead D, Tsang YW, Rajpoot N (2019) Mild-net: minimal information loss dilated network for gland instance segmentation in colon histology images. Med Image Anal 52:199–211
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Kamnitsas K, Ledig C, Newcombe VF, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B (2017) Efficient multi-scale 3d cnn with fully connected crf for accurate brain lesion segmentation. Med Image Anal 36:61–78
Kim M, Yun J, Cho Y, Shin K, Jang R, Hj Bae, Kim N (2020) Deep learning in medical imaging. Neurospine 17(2):471
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436 . https://doi.org/10.1038/nature14539
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, Van Der Laak JA, Van Ginneken B, Sánchez CI (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88
Lundervold AS, Lundervold A (2019) An overview of deep learning in medical imaging focusing on mri. Zeitschrift Für Medizinische Physik 29 (2):102–127
Ponzio F, Macii E, Ficarra E, Di Cataldo S (2018) Colorectal cancer classification using deep convolutional networks. In: Proceedings of the 11th international joint conference on biomedical engineering systems and technologies, vol 2, pp 58–66
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv:14091556
Sudana O, Gunaya IW, Putra IKGD (2020) Handwriting identification using deep convolutional neural network method. Telkomnika 18(4):1934–1941
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1–9
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2818–2826
Torrey L, Shavlik J (2010) Transfer learning. In: Handbook of research on machine learning applications and trends: algorithms, methods, and techniques. IGI Global, pp 242–264
Tou JT, Gonzalez RC (1974) Pattern recognition principles. prp
Wu N, Phang J, Park J, Shen Y, Huang Z, Zorin M, Jastrzkebski S, Févry T, Katsnelson J, Kim E, et al. (2019) Deep neural networks improve radiologists’ performance in breast cancer screening. IEEE Transactions on Medical Imaging
Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A (2016) Learning deep features for discriminative localization. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2921–2929
Acknowledgements
This work was supported by the GRRC program of Gyeonggi province. [GRRC KGU 2020-B04, Image/Network-based Intellectual Information Manufacturing Service Research]
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sang-Hyun Kim and Hyun Min Koh have contributed equally in this article as first authors.
Rights and permissions
About this article
Cite this article
Kim, SH., Koh, H.M. & Lee, BD. Classification of colorectal cancer in histological images using deep neural networks: an investigation. Multimed Tools Appl 80, 35941–35953 (2021). https://doi.org/10.1007/s11042-021-10551-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-021-10551-6