Abstract
In order to detect the cerebral microbleed (CMB) voxels within brain, we used susceptibility weighted imaging to scan the subjects. Then, we used undersampling to solve the accuracy paradox caused from the imbalanced data between CMB voxels and non-CMB voxels. we developed a seven-layer deep neural network (DNN), which includes one input layer, four sparse autoencoder layers, one softmax layer, and one output layer. Our simulation showed this method achieved a sensitivity of 95.13%, a specificity of 93.33%, and an accuracy of 94.23%. The result is better than three state-of-the-art approaches.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Cerebral microbleed (CMB) [49] is small foci of chronic blood products in normal brain tissues. They are closely related with glomerular filtration [35], dementia [55], cortical superficial siderosis [26], and ageing [16]. They are important recognized entity with the rapid development of magnetic resonance imaging (MRI) especially the susceptibility weighted imaging (SWI). The hemosiderin within CMB foci is superparamagnetic, which causes significant local inhomogeneity in the magnetic field around CMB, leading fast decay of MRI signal. Hence, CMB appear hypointensity in the scanned image.
Traditional interpretation depends on the MARS (microbleed anatomical rating scale) [22] that draws up stringent rules to classify CMB into two types: “definite” and “possible” [3]. Nevertheless, the manual interpretation are not reliable due to the high intra-observer and inter-observer variability. Visual screening is prone to either confuse with CMB mimics or miss small CMBs [46].
In the last decade, computer scientists tried to solve this problem based on computer vision and image processing techniques. Fazlollahi (2015) [20] combined multi-scale mechanism and Laplacian of Gaussian approach. They abbreviated it as MSLoG. They also used random forest (abbreviated as RF) classifiers. Seghier (2011) [54] proposed a microbleed detection via automated segmentation (MIDAS) technique. Barnes (2011) [4] relied on a statistical thresholding algorithm to detect the hypointensity. They then used support vector machine (SVM) classifier to separate true CMB from others. Bian (2013) [6] employed a 2D fast RST to detect putative CMBs. Afterwards, false results were removed using features of geometry. Kuijf (2012) [27] presented a radial symmetry transform (RST) method. Charidimou (2012) [8] discussed the principles, methodologies, and rational of CMB and its mapping in vascular dementia. Bai (2013) [2] detected CMBs in super-acute ischemic stroke patients treated with intravenous thrombolysis. Roy (2015) [52] proposed a novel multiple radial symmetry transform (MRST) and RF method. Chen (2016) [9] used leaky rectified linear unit (LReLU). Hou (2016) [24] proposed a four-layer deep neural network (DNN) method.
Nevertheless, the detection accuracy of above methods are still quite low. For example: Bai’s method [2] combined multi-modality imaging, but they did not use computer vision approach to increase the identification performance. Roy’s method [52] obtained a sensitivity of 85.7%, which is quite higher than human interpretation, but it did not explore the power of computer vision fully. Chen’s method [9] validated that LReLU performed better than other activation functions, but that study lacks theoretical analysis. Hou’s method [24] showed DNN has better result, but the structure of their DNN is shallow, which did not explore the powerfulness of DNN.
Recently, the “deep learning” technique [19] has been proposed for machine learning. It gained burning interests and achieved remarkable achievements. The AlphaGo [10] just used deep learning to beat the world champion in five-game match. It is the 1st time that a computer machine beats a 9-dan professional [56]. Besides, deep learning has been successfully applied in system identification [51], human activity recognition [50], video tracking [60], facial retouching detection [5], etc.
In this study, we aimed to use the deep learning technique to realize the CMB detection. We chose to use the sparse autoencoder (SAE) and softmax classifier. The structure of remainder is organized as follows: Section 2 gives the details of subjects. Section 3 presents the methodology. Section 4 offers the results and discussions. Finally, Section 5 concludes the paper.
2 Subjects
Ten cerebral autosomal-dominant arteriopathy with subcortical infarcts and Leukoencephalopathy (shorted as CADASIL) patients and ten healthy controls (HCs) were enrolled. We reconstructed the 3D volumetric image by Syngo MR B17 software. The size of each subject is the same as 364x448x48.
Three neuroradiologists with over twenty years of experiences carried out manual detection of CMBs. The labelled “possible” and “definite” were all regarded as CMB voxels, and others were regarded as non-CMB voxels. CMB voxels are shown within the red curve in Fig. 1. The exclusion criteria contains two rules: (1) blood vessels were discarded by tracking through neighboring slices; (2) lesions larger than 10 mm were not considered.
Slice of cerebral microbleed, SI represents slice index (The SWIs were scanned by 3 T SIEMENS Verio scanner with station of MRC 40810. Slice number = 48, sequence = swi3d1r, flip angle =15 degree, The bit depth = 12, resolution = [0.5 × 0.5 × 2]mm3, slice thickness = 2 mm, echo time = 20 ms, repetition time = 28 ms, bandwidth =120 Hx/px)
2.1 Dataset generation
Sliding neighborhood processing (SNP) technique was employed to generate the input and target datasets from the 20 volumetric 3D brain images. We process on each slice of each subject. As we know, the neighborhood of a pixel p is a matrix, we vectorize this matrix to form a input sample x, then the status of the central pixel is defined as its target value y. Mathematically,
where N represents the neighborhood and V represents the vectorization operation. The final input dataset X and target dataset Y are formed by processing all voxels in set A.
Here A represents the voxels of all slices of all subjects except the border.
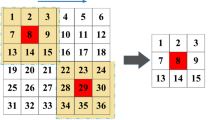
In this study, we choose the window size of 61 × 61 pixels, namely, the voxels of 30-pixel borders are discarded as shown in Fig. 2. The window moves towards right and down so as to cover the set A. Finally, we generated 68,847 CMB voxels and 113,165,073 non-CMB voxels.
2.2 Accuracy paradox
The imbalanced data will cause severe problem to the classification [30, 41, 43], since now the non-CMB voxels are 1644 times of CMB voxels. The classifier is prone to be trained nonsense as output 1 always. This will give the performance in Table 1. The sensitivity is 0%, the specificity is 100.00%, and the accuracy is 99.93%. This suggests us the specificity and accuracy are not a good indicator in this study. Therefore, we will focus more weight on the sensitivity measure.
This imbalanced data problem arise from the area of foci of microbleed is extremely small compared to healthy tissues. This causes the “accuracy paradox [59]” as shown in Table 1. Many methods can solve or mitigate the imbalanced data problem, such as cost function based techniques [25] and sampling based approaches [15, 21]. In this study, we use the undersampling technique [1] to reduce the 113,165,073 to 68,854 samples.
3 Methodology
Artificial intelligence has developed four phases in medicine. Originally, expert system or knowledge-based systems write hard-code knowledge or rules in their programs. Later, traditional machine learning approaches used handcrafted features: (1) the physical features, for example, the cortical thickness, the area of some specific brain tissue; (2) the mathematical features, for example, wavelet transform [37], wavelet entropy [33], contourlet transform [23], fractional Fourier transform [7, 28, 57], gray-level co-occurrence matrix [45], eigenvector [47], and etc.
In the last decade, representation learning (RL) aimed to learn features from data. Its goal is to discover low-dimensional features, which can capture the structure of the input high-dimensional brain images. Several years ago, the deep learning was proposed to learn simple and abstract features by multiple layers. The four phases are depicted in a Venn diagram shown in Fig. 3. Especially, the latest deep learning techniques have shown successful application in a massive of fields. Li (2016) [31] used deep neural network in underwater image descattering. Morabito (2017) [44] employed deep learning representation to detect early-stage Creutzfeldt-Jakob disease. Tabar (2017) [58] used deep learning approach to classify EEG motor imagery signals. All these methods have shown the superiority of deep learning to traditional schools of artificial intelligence.
Currently, there are too types of mature deep learning techniques. One is the convolutional neural network (CNN). The other is the autoencoder. The CNN was inspired by animal visual cortex, but the overfitting may happen when applying complicated full-connected layers [32]. The autoencoder is famous for its learning generative models of data. Besides, it is easy to create its model and to train it. In this study, we choose the sparse auto-encoder and softmax classifier.
3.1 Autoencoder
Autoencoder is a symmetrical neural network that learns the features in an unsupervised manner. The autoencoder is successfully applied in image reconstruction [42], image super-resolution [63], prediction [53], etc. The structure of AE is shown in Fig. 4, where the encoder part is with weight E = [e(1), e(2), …, e(m)] and bias B 1 = [b 1(1), b 1(2), … b 1(m)], the decoder part is with weight D = [d(1), d(2), …, d(m)] and bias B 2 = [b 2(1), b 2(2), … b 2(m)]. The encoder and decoder parts combined and make the output data Y = [y 1, y 2, …, y n ] to be equal to input vector X = [x 1, x 2, …, x n ]. Suppose the activation function is logistic sigmoid form, we have
where A = [a 1, a 2, …, a m ] is the output of hidden layer. Then, the decoding of A is carried out as
3.2 Sparse autoencoder
To minimize the error between the input vector X and output Y, we can yield the objective function as
From eq. (5)(6), we can deduce Y can be expressed as
Hence, eq. (7) can be revised as
To avoid over-complete mapping or learn a trivial mapping, we add one regularization term on the weight and one regularization term of a sparse constraint:
where α is the weight of sparse penalty, and β the regularization factor controlling the degree of weight decay. K() is the Kullback-Leibler divergence defined as
The symbol ρ represents the desired probability of being activated, and ρ j the average activation probability of j-th hidden neuron. The training procedure is performed by scaled conjugate gradient descent (SCGD) method.
3.3 Softmax classifier
The softmax classifier is put as the last layer in the deep neural network, aiming to classifying the learned features from sparse autoencoders beforehand. Remember that a logistic regression is a binary classifier with definition as:
where θ represents the model parameters.
In contrast, the softmax classifier use softmax as the activation function and it can be regarded as a multinomial logistic regression with output has k values as:
The values of parameters θ can be obtained by iterative optimization algorithm on the loss function, which used cross entropy in this study. The softmax classifier can be regarded as the multinomial logistic regression [64].
3.4 Deep neural network structure
The SAE was stacked to extract brain image features gradually. The feature code of each hidden layer was transmitted to next layer, as shown in Fig. 5.
The structure of the proposed deep neural network (DNN) was established in Fig. 5. Here we create a seven-layer DNN, consisting of one input layer, four SAE layers, one softmax layer, and one output layer. The four SAE layers share the same structure, but their sizes are different. The size of each layer was selected by experience:
-
The input layer has 61*61 = 3721 neurons;
-
The first SAE layer has 1500 hidden neurons;
-
The second SAE layer has 900 hidden neurons;
-
The third SAE layer has 500 hidden neurons;
-
The fourth SAE layer has 100 hidden neurons;
-
The softmax has one neuron indicates CMB voxel or non-CMB voxel;
-
The output layer is directly linked to the softmax layer.
In total, we create a seven-layer DNN with structure of 3721–1500–900-500-100-1-1. Remember weights and biases are assigned to only the SAE and softmax layers, they are not assigned to the input and output layer. For statistical analysis, 10-fold cross validation [65] was used, and the average out-of-sample error was reported. The SAE is reported to have better performance than support vector machine (SVM) [17] and its variants: the fuzzy SVM [61], generalized eigenvalue proximal SVM [34], and twin SVM [62].
4 Results and discussions
The program was developed in-house via the Neural Network Toolbox in Matlab R2016a. We used the functions of the built-in “autoencoder” class. The programs were run on the IBM laptop with 3.2GHz i5–3470 CPU, 4GB RAM, and Windows 10 operating system.
4.1 Sliding neighborhood
The sliding neighborhood technique extracts the neighborhood of central voxel as the input of each sample, and the status of that central voxel as the target. Figure 6 shows nine examples of CMB voxels and nine examples of non-CMB voxels. We can observe that the 61 × 61 neighborhood is large enough for the human interpretation, thus, the window size is reasonable.
4.2 10-fold segmentation
We divide the 137,701 samples into 10 folds at random. The detailed results are shown in Table 2. Here the sum of samples in each fold is equal to 137,701, for all 10 runs. This segmentation meets the requirement of stratification, i.e., the class distribution at each fold are almost the same. Take the first row as an example, it means the first fold contains 13,771 samples (6885 CMB samples and 6886 non-CMB samples), the second fold contains 13,770 samples (6885 CMB samples and the same-size non-CMB samples), the third fold also contains 13,770 samples with each class of 6885, the fourth fold contains 13,770 samples (6884 CMB samples and 6886 non CMB-samples), and the fifth to tenth fold contains 13,770, 13,771, 13,770, 13,770, 13,770, 13,769 samples, respectively.
4.3 Identification result
We report the 10 × 10-fold cross validation identification result of our seven-layer deep neural network in Table 3. Taking the first run as an example, our algorithm identifies [6406 6346 6450 5975 6749 6422 6417 6777 6804 6718] CMB voxels and [6841 6182 6082 6565 6617 6371 6129 5977 6632 6450] non-CMB voxels correctly over the ten folds. Summarizing both CMB voxels and non-CMB voxels, we identify [13,247 12,528 12,532 12,540 13,366 12,793 12,546 12,754 13,436 13,168] voxels correctly over ten folds. In ten folds, we identified correctly 65,064 CMB voxels and 63,846 non-CMB voxels at the first run.
Some latest feature extraction methods may increase the identification performance, such as: curve structure [13, 14], Zernike moment [18], fractional dimension [11], etc. Furthermore, some traditional classifiers, for example, extreme learning machine [36], linear regression classifier [12], and Bayesian classifier [48] will be taken as competing classifiers in the future studies.
(R.I. Run Index, F.I. Fold Index, a + b = c represents a samples correctly identified as CMB voxels and b samples correctly identified as non-CMB voxels. In total, c samples are identified correctly.
4.4 Measures of classification performance
The classification performance of our method over 10 runs of 10-fold cross validation is shown in Table 4. On average, the sensitivity is 95.13 ± 0.84%, the specificity is 93.33 ± 0.84%, and the accuracy is 94.23 ± 0.84%. The sensitivity is the most important measure, since it can detect the CMB from healthy control. The specificity is less important, since misclassification of healthy people can be corrected in further diagnosis.
4.5 Comparison to state-of-the-art
Finally, we compare this 7-layer SAE-DNN method with MRST + RF [52], LReLU [9], and 4-layer DNN [24]. The comparison results in Table 5 and Fig. 7 showed that our method gives better results in sensitivity and accuracy. The MRST + RF [52] method gives the highest specificity. In all, our method is better than both MRST + RF [52] and 4-layer DNN [24] in terms of sensitivity and accuracy.
Our specificity result of 93.33% is lower than MRST + RF [52] of 99.5%. Nevertheless, in clinical condition, the sensitivity (i.e., to identify CMB voxel) is the most important. The low specificity (i.e., to identify non-CMB voxel) can be second-checked by human neuroradiologists in a fast way. In the future, we shall test convolutional neural network [38]. We shall also try to generalize our method to real-time visual system [29].
A shortcoming of our method is that for SWI images from two scanners with different setting, the contrast of gray-level image may differ. To solve the problem, we may need to use “image enhancement [39]” or “light compensation [40]” techniques.
5 Conclusions
In this study, our team proposed a new 7-layer SAE based deep neural network for cerebral microbleed detection. The results showed that this method is promising and gives better results than three state-of-the-art methods: MRST + RF [52], LReLU [9], and 4-layer DNN [24].
In the future, we shall enroll more subjects to increase the reliability and robustness of our method. Besides, we shall test other advanced classifiers, such as linear regression classifier, extreme learning machine, etc.
References
Anand A et al (2010) An approach for classification of highly imbalanced data using weighting and undersampling. Amino Acids 39(5):1385–1391
Bai QK et al (2013) Susceptibility-weighted imaging for cerebral microbleed detection in super-acute ischemic stroke patients treated with intravenous thrombolysis. Neurol Res 35(6):586–593
Banerjee G et al (2016) Impaired renal function is related to deep and mixed, but not strictly lobar cerebral microbleeds in patients with ischaemic stroke and TIA. J Neurol 263(4):760–764
Barnes SRS et al (2011) Semiautomated detection of cerebral microbleeds in magnetic resonance images. Magn Reson Imaging 29(6):844–852
Bharati A et al (2016) Detecting facial retouching using supervised deep learning. IEEE Transactions on Information Forensics and Security 11(9):1903–1913
Bian W et al (2013) Computer-aided detection of radiation-induced cerebral microbleeds on susceptibility-weighted MR images. Neuroimage Clin 2:282–290
Cattani C, Rao R (2016) Tea category identification using a novel fractional Fourier entropy and Jaya algorithm. Entropy 18(3):77
Charidimou A et al (2012) Cerebral microbleed detection and mapping: principles, methodological aspects and rationale in vascular dementia. Exp Gerontol 47(11):843–852
Chen Y (2016a) Voxelwise detection of cerebral microbleed in CADASIL patients by leaky rectified linear unit and early stopping: a class-imbalanced susceptibility-weighted imaging data study. Multimedia Tools and Applications. doi:10.1007/s11042-017-4383-9
Chen JX (2016b) The evolution of computing: AlphaGo. Computing in Science & Engineering 18(4):4–7
Chen X-Q (2016c) Fractal dimension estimation for developing pathological brain detection system based on Minkowski-Bouligand method. IEEE Access 4:5937–5947
Chen Y (2017) A feature-free 30-disease pathological brain detection system by linear regression classifier. CNS Neurol Disord Drug Targets 16(1):5–10
Chen Y et al (2016a) Curve-like structure extraction using minimal path propagation with backtracking. IEEE Trans Image Process 25(2):988–1003
Chen Y et al (2016b) Structure-adaptive fuzzy estimation for random-valued impulse noise suppression. IEEE Trans Circuits Syst Video Technol. doi:10.1109/TCSVT.2016.2615444
D'Addabbo A, Maglietta R (2015) Parallel selective sampling method for imbalanced and large data classification. Pattern Recogn Lett 62:61–67
Del Brutto OH et al (2016) Oily fish consumption is inversely correlated with cerebral microbleeds in community-dwelling older adults: results from the Atahualpa project. Aging Clin Exp Res 28(4):737–743
Dong Z (2014) Classification of Alzheimer disease based on structural magnetic resonance imaging by kernel support vector machine decision tree. Prog Electromagn Res 144:171–184
Du S (2017) Alzheimer's disease detection by pseudo Zernike moment and linear regression classification. CNS Neurol Disord Drug Targets 16(1):11–15
Erfani SM et al (2016) High-dimensional and large-scale anomaly detection using a linear one-class SVM with deep learning. Pattern Recogn 58:121–134
Fazlollahi A et al (2015) Computer-aided detection of cerebral microbleeds in susceptibility-weighted imaging. Comput Med Imaging Graph 46:269–276
Fithian W, Hastie T (2014) Local case-control sampling: efficient subsampling in imbalanced data sets. Ann Stat 42(5):1693–1724
Gregoire SM et al (2009) The microbleed anatomical rating scale (MARS) reliability of a tool to map brain microbleeds. Neurology 73(21):1759–1766
Heshmati A et al (2016) Scheme for unsupervised colour-texture image segmentation using neutrosophic set and non-subsampled contourlet transform. IET Image Process 10(6):464–473
Hou X-X, Chen H (2016) Sparse autoencoder based deep neural network for voxelwise detection of cerebral microbleed. In 22nd International Conference on Parallel and Distributed Systems. Wuhan: IEEE. pp. 1229–1232
Hwang JP et al (2011) A new weighted approach to imbalanced data classification problem via support vector machine with quadratic cost function. Expert Syst Appl 38(7):8580–8585
Inoue Y et al (2016) Diagnostic significance of cortical superficial siderosis for Alzheimer disease in patients with cognitive impairment. Am J Neuroradiol 37(2):223–227
Kuijf HJ et al (2012) Efficient detection of cerebral microbleeds on 7.0 T MR images using the radial symmetry transform. NeuroImage 59(3):2266–2273
Li J (2016) Detection of left-sided and right-sided hearing loss via fractional Fourier transform. Entropy 18(5):194
Li YJ et al (2014) Real-time visualization system for Deep-Sea surveying. Mathematical Problems In Engineering. doi:10.1155/2014/437071
Li Y et al (2016a) Grouped variable selection using area under the ROC with imbalanced data. Communications in Statistics-Simulation and Computation 45(4):1268–1280
Li YJ et al (2016b) Underwater image de-scattering and classification by deep neural network. Comput Electr Eng 54:68–77
Li H et al (2017) Vehicle detection in remote sensing images using denoizing-based convolutional neural networks. Remote Sensing Letters 8(3):262–270
Liu G (2015a) Pathological brain detection in MRI scanning by wavelet packet Tsallis entropy and fuzzy support vector machine. Springerplus 4(1):716
Liu A (2015b) Magnetic resonance brain image classification via stationary wavelet transform and generalized eigenvalue proximal support vector machine. Journal of Medical Imaging and Health Informatics 5(7):1395–1403
Liu YY et al (2016) Association between low estimated glomerular filtration rate and risk of cerebral small-vessel diseases: a meta-analysis. J Stroke Cerebrovasc Dis 25(3):710–716
Lu S, Qiu X (2017) A pathological brain detection system based on extreme learning machine optimized by bat algorithm. CNS Neurol Disord Drug Targets 16(1):23–29
Lu HM et al (2012) Maximum local energy: an effective approach for multisensor image fusion in beyond wavelet transform domain. Computers & Mathematics with Applications 64(5):996–1003
Lu H et al (2016a) Wound intensity correction and segmentation with convolutional neural networks. Concurrency and Computation: Practice and Experience. doi:10.1002/cpe.3927
Lu HM et al (2016b) Underwater image enhancement method using weighted guided trigonometric filtering and artificial light correction. J Vis Commun Image Represent 38:504–516
Lu HM et al (2016c) Turbidity underwater image restoration using spectral properties and light compensation. IEICE Trans Inf Syst E99D(1):219–227
Mao WT et al (2016) Two-stage hybrid extreme learning machine for sequential imbalanced data. In International Conference on Extreme Learning Machine (ELM). Hangzhou: Springer Int Publishing Ag. pp. 423–433
Mehta J, Majumdar A (2017) RODEO: robust DE-aliasing autoencOder for real-time medical image reconstruction. Pattern Recogn 63:499–510
Mirza B, Lin ZP (2016) Meta-cognitive online sequential extreme learning machine for imbalanced and concept-drifting data classification. Neural Netw 80:79–94
Morabito FC et al (2017) Deep learning representation from electroencephalography of Early-Stage Creutzfeldt-Jakob disease and features for differentiation from rapidly progressive dementia. In J Neural Syst 27(2):15 Article ID: 1650039
Pantic I et al (2016) Fractal analysis and gray level co-occurrence matrix method for evaluation of reperfusion injury in kidney medulla. J Theor Biol 397:61–67
Peng Q et al (2016) Longitudinal relationship between chronic kidney disease and distribution of cerebral microbleeds in patients with ischemic stroke. J Neurol Sci 362:1–6
Phillips P (2016) Three-dimensional Eigenbrain for the detection of subjects and brain regions related with Alzheimer's disease. J Alzheimers Dis 50(4):1163–1179
Rajaguru H, Prabhakar SK (2016) A framework for epilepsy classification using modified sparse representation classifiers and naive Bayesian classifier from electroencephalogram signals. Journal of Medical Imaging and Health Informatics 6(8):1829–1837
Romero JR et al (2012) Lipoprotein phospholipase A2 and cerebral microbleeds in the Framingham heart study. Stroke 43(11):3091–U525
Ronao CA, Cho SB (2016) Human activity recognition with smartphone sensors using deep learning neural networks. Expert Syst Appl 59:235–244
de la Rosa E, Yu W (2016) Randomized algorithms for nonlinear system identification with deep learning modification. Inf Sci 364:197–212
Roy S et al (2015) Cerebral microbleed segmentation from susceptibility weighted images. Proc SPIE 9413:94131E
Saha M et al (2016) Autoencoder-based identification of predictors of Indian monsoon. Meteorog Atmos Phys 128(5):613–628
Seghier ML et al (2011) Microbleed detection using automated segmentation (MIDAS): a new method applicable to standard clinical MR images. PLoS One 6(3):e17547
Shams S et al (2015) Cerebral microbleeds: different prevalence, topography, and risk factors depending on dementia diagnosis-the Karolinska imaging dementia study. Am J Neuroradiol 36(4):661–666
Silver D et al (2016) Mastering the game of go with deep neural networks and tree search. Nature 529(7587):484
Sun Y (2016) A multilayer perceptron based smart pathological brain detection system by fractional Fourier entropy. J Med Syst 40(7):173
Tabar YR, Halici U (2017) A novel deep learning approach for classification of EEG motor imagery signals. J Neural Eng 14(1):11 Article ID: 016003
Valverde-Albacete FJ, Pelaez-Moreno C (2014) 100% classification accuracy considered harmful: the normalized information transfer factor explains the accuracy paradox. PLoS One 9(1):e84217
Xue HY et al (2016) Tracking people in RGBD videos using deep learning and motion clues. Neurocomputing 204:70–76
Yang J (2015) Identification of green, oolong and black teas in China via wavelet packet entropy and fuzzy support vector machine. Entropy 17(10):6663–6682
Yang M (2016) Dual-tree complex wavelet transform and twin support vector machine for pathological brain detection. Appl Sci 6(6):169
Zeng K et al (2017) Coupled deep autoencoder for single image super-resolution. IEEE Transactions on Cybernetics 47(1):27–37
Zhan TM, Chen Y (2016) Multiple sclerosis detection based on biorthogonal wavelet transform, RBF kernel principal component analysis, and logistic regression. IEEE Access 4:7567–7576
Zhang YD et al (2016) Facial emotion recognition based on biorthogonal wavelet entropy, fuzzy support vector machine, and stratified cross validation. IEEE Access 4:8375–8385
Acknowledgements
This paper was supported by NSFC (61602250), Leading Initiative for Excellent Young Researcher (LEADER) of Ministry of Education, Culture, Sports, Science and Technology-Japan (16809746), Natural Science Foundation of Jiangsu Province (BK20150983), Program of Natural Science Research of Jiangsu Higher Education Institutions (16KJB520025), Open Research Fund of Hunan Provincial Key Laboratory of Network Investigational Technology (2016WLZC013), Open Fund of Fujian Provincial Key Laboratory of Data Intensive Computing (BD201607), Open fund for Jiangsu Key Laboratory of Advanced Manufacturing Technology (HGAMTL1601), Open fund of Key Laboratory of Guangxi High Schools Complex System and Computational Intelligence (2016CSCI01).
Author information
Authors and Affiliations
Corresponding author
Additional information
Yu-Dong Zhang, Yin Zhang, Xiao-Xia Hou and Hong Chen contributed equally to this paper
Rights and permissions
About this article
Cite this article
Zhang, YD., Zhang, Y., Hou, XX. et al. Seven-layer deep neural network based on sparse autoencoder for voxelwise detection of cerebral microbleed. Multimed Tools Appl 77, 10521–10538 (2018). https://doi.org/10.1007/s11042-017-4554-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-017-4554-8