Abstract
Medical image registration is commonly used in clinical diagnosis, treatment, quality assurance, evaluation of curative efficacy and so on. In this paper, the edges of the original reference and floating images are detected by the B-spline gradient operator and then the binarization images are acquired. By computing the binarization image moments, the centroids are obtained. Also, according to the binarization image coordinates, the rotation angles of the reference and floating images are computed respectively, on the foundation of which the initial values for registering the images are produced. When searching the optimal geometric transformation parameters, the modified peak signal-to-noise ratio (MPSNR) is viewed as the similarity metric between the reference and floating images. At the same time, the simplex method is chosen as multi-parameter optimization one. The experimental results show that, this proposed method has a fairly simple implementation, a low computational load, a fast registration and good registration accuracy. It also can effectively avoid trapping in the local optimum and is adapted to both mono-modality and multi-modality image registrations. Also, the improved iterative closest point algorithm based on acquiring the initial values for registration from the least square method (LICP) is introduced. The experiments reveal that the measure acquiring the initial values for registration from image moments and the least square method (LSM) is feasible and resultful strategy.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Along with the wide and in-depth applications of medical imaging technologies, to clinical medicine such as disease diagnosis, surgical exploration, radiation therapy, assessment of healing efficacy and so on, the research about registering the medical images generated by various imaging devices, becomes a hot field which has been drawing more and more attention over the years.
Medical image registration, as a important image processing technology, denotes that the use of the spatial geometric transformation to align two or more images sampling from a different time, from a different scene or from a different modality, enables the pixels (voxels) representing the same structure to achieve the space correspondence [5,12,13,15,16,23,30]. Over the past several decades, medical image registration methods have made great progress, and researchers around the world have introduced many practical methods, which are divided into two broad categories: feature-based and intensity-based medical image registration methods [17]. As for the former, by searching the common, obvious and significant features between the registering images, it obtains the optimal transformation parameters. This is a simple and fast method, but its registration accuracy heavily depends on whether the image feature points can be exactly extracted [6,11]. Due to the complexity of medical images, it is difficult to automatically extract the useful feature points; thus it is necessary to interactively pick out them. Therefore, its poor adaptability needs to be further improved. With respect to the intensity-based image registration, it needn’t capture the feature points and only uses the gray level similarity between two images as the registration metric. Due to the full use of image gray information, it has fairly higher registration accuracy. Among intensity-based methods published, the mutual information (MI) technology has become one of the most prevalent methods because it is more flexible and accurate than any other methods in the global information context [6,11,17,21]. In the process of the image registration, it employs MI as similarity metric, and can automatically, directly and accurately align two or more images without any prior segmentation or other preprocessing, which makes it to be popular and to be very suitable for clinical applications. In the literature [20], an alignment method maximizing mutual information of medical images was proposed to obtain the parameters for the translation and rotation needed to register by Paul et al. Since the mutual information function contains many local maxima, it easily gets into the local optimum. Therefore, the transformation parameters are not the global optimum ones. Fei et al. [8], Slomka et al. [26] and Radau et al. [22], by using multi-parameters optimizations of Powell and simplex methods, obtained the parameters for the translation and rotation. The MI technology, however, has the following inherent limitations. First, due to the fact that it only considers the gray information instead of or regardless of spatial information, it often traps in the local optimum and even fails to register images. Second, although MI reflects the information of the overlapping region between two images, it cannot directly embody the gray discrepancy between two registering images. That is, it cannot be reckoned as the criterion for objectively judging the quality of the registered image. In addition, it has a heavily computational load, time-consuming process and low registration efficiency. Because of these reasons above, Pan et al. [18] proposed medical image registration based on singular value decomposition and modified peak signal-to-noise ratio. In this paper, the medical image moments are calculated to get the centroid, and the rotation angles of the reference and floating images are computed respectively based on the rotational invariance of the singular values of the matrix of the medical image coordinates, on the basis of which the initial values for registering the images are produced. When searching the optimal geometric transformation parameters, the modified peak signal-to-noise ratio is selected as the similarity measure, and the simplex method as multi-parameter optimization. The experimental results show that, this proposed method is fairly simple to implement and has a low computation load and good registration accuracy. It also can effectively avoid trapping in the local optimum.
Inspired by [18], in order to address the above problems, on the basis of a comprehensive and thorough research into image moments, the least square method (LSM) and peak signal-to-noise ratio (PSNR), we introduce another registration method, i.e. medical image registration based on LSM and modified PSNR (MPSNR). In this paper, the binarization images are acquired after the B-spline gradient operator (BSGO) is used to detect the edges of the reference and floating images. Based on computing the moments of the binarization images, the centroids are obtained respectively, and then according to the binarization image coordinates and LSM, the rotation angles of the reference and floating images are calculated respectively, on the foundation of which the initial values for registration are acquired. MPSNR, as a similarity metric between two images, incorporating with the simplex method, is applied to explore the optimal transformation parameters. This proposed method can cater to both mono-modality and multi-modality image registrations. In addition, the method acquiring the initial values from image moments and LSM is adopted for the iterative closest point (ICP) algorithm, and also can advance the performance of ICP.
2 Imperfection of the image registration based on MI
Suppose that reference image R and floating image F are both of M × N pixels with the upper left pixel being (1,1) and the gray level being represented by L, and the gray values at point (x, y) are r (x, y) and f (x, y) respectively. For any image I, its entropy is given by the expression [15]
where p(k) represents the probability function that gray value k appears.
MI, as an important concept in the field of information theory, is a prevalent entropy-based similarity metric extensively used in medical image registration. This metric is superior to most other intensity-based ones in multi-modality registrations because it only assumes a statistical dependence between two images. MI is a measure of the degree of dependence of two images. In the process of aligning two or more images, when each point in the floating image is mapped onto its corresponding point in the reference image, registration is achieved by maximizing the gray information, that is, in this case, the information that the floating image can represent the reference image is the richest, which is so-called MI expressed by:
where H(R) and H(F), are the entropies of reference image R and floating image F respectively, and H(R, F) is the joint entropy of R and F. Summarily speaking, the MI-based registration (MIR) is described:
That is, finding an optimal transformation T optimal maximizes the MI. Generally, 2-D medical image rigid registration, in essence, includes translation and rotation transformations, namely
where ∆x and ∆y, express vertical and horizontal displacements respectively, and ϕ denotes the rotation angle. From this, exploring the optimal transformation T optimal is essentially a multi-parameter optimization process as well.
Among the methods of solving the multi-parameter optimization, simplex, pattern search and Powell methods, as unconstrained optimization ones, are commonly used for registering two images [4]. In the literatures [8,20,22,26], by the selection of the MI as similarity metric, authors adopted Powell, pattern search, and simplex methods to get the translation and rotation transformation parameters. Since involving a lot of local optima in the MI function, these methods mentioned above easily get into the local optima. In the process of seeking the optimal solution, these methods have to recursively and iteratively explore the solution space, and consequently their computational loads are very heavy. Moreover, whether these optimization methods successfully can get the optimal parameters strongly depends on the initial values selected. If they are ill-suited, then the registration process needs more searching time and even fail to register the images.
3 Medical image registration based on LSM and MPSNR
In order to advance registration efficiency and reduce the possibility of failing to register images, this proposed method first acquires the initial values for registering the images to curtail the time for searching the parameter space and to avoid getting into the local optimum, and then selecting MPSNR as similarity metric further reduces the time for registering the images.
3.1 Edge detection and extraction of feature points
B-spline, as a curve and surface fitting function in computer graphics, is used increasingly in the field of image processing. Unser [31] comprehensively and thoroughly investigated into the combination with the spline functions and image processing, and the applications of the B-spline function were discussed in detail. Hereafter, many new methods were proposed to be adopted for image processing, such as a spline-based image registration [29], a fast parametric elastic image registration based on B-spline function [13], a 3-D medical image reconstruction based on B-spline function interpolation [3], a new medical image registration based on B-spline rigid-elastic transformation [28], a color image segmentation based on B-spline model [25], a magnetic resonance image segmentation based on B-spline snakes [27] and so on.
In this paper, in order to get the rotation angle of and extract the feature points from an original image, we propose an edge detection method based on cubic B-spline function.
Definition 1 The B-spline of order n is defined as follows[32,33]:
where \( \mu (x) = \left\{ {\matrix{ {0,} \hfill &{x << 0} \hfill \\ {1,} \hfill &{x \geqslant 0} \hfill \\ }<!end array> } \right. \) is the unit step function, and \( C_{{n + 1}}^j \) is a combinatorial formula. According to the properties of \( {{B}_n}(x) \) [32], it satisfies the requirement of the smoothing function. As introduced in the literatures [32,33], the smoothness and approximation of a smoothing function are mutually contradictory. In essence, the smoothness of \( {{B}_n}(x) \) is equal to a low-pass filter. With n increasing, the smoothing performance of \( {{B}_n}(x) \) is strengthened and its filtering effect is improved as well, but its processing time correspondingly increases. Conversely, when n reduces, the approximation performance of \( {{B}_n}(x) \) is improved, its edge protection ability is enhanced and the details are well retained, but the low-pass effect decreases. In this paper, in order to counterbalance the smoothness and the approximation, let n = 3, namely, cubic B-spline function \( {{B}_3}(x) \), which is defined by Eq. (6) as follows:
The medical image f(x,y) can be regarded as a uniform sampling for a surface; therefore we use the B-spline function to smooth it. The medical image is approximately expressed by a surface as follows [14]:
From Eqs. (7) and (8), the surface S(x,y) is referred to as the result from the discrete convolution among the image f(x,y), k-order and l-order B-spline functions. Here, let \( k = l = 3 \). When x and y are integers, \( {{N}_{{k,l}}}\left( {x,y} \right) \) is the 3 × 3 neighborhood around the pixel (x, y). From Eq. (7), the surface S(x,y) has second-order continuous differentiability, that is, S(x,y) is a smooth one. According to the local properties of B-spline function, Eqs. (7) and (8), the following Eq. (9) is deduced:
From Eq. (9), the x and y directional derivative are shown as follow:
By Eqs. (9), (10) and (11), we can calculate the directional derivative convolution template along x and y of the surface fitting by cubic B-spline function respectively:
A gradient operator is often used to detect potential object boundaries or edges. The gradient operator applied to a continuous function generates a vector at each point whose direction indicates the direction of the maximum change of the function at that point, and whose magnitude denotes the magnitude of this maximum change. For an image, its edge generally lies in the sudden change areas of the gray values, and the gradient is most commonly-used differential method in the image edge detection, namely the sudden change point of signal is in correspondence with the maximum module value of the first derivative. Regarding the image F, the gradient at point (x, y) is defined as the vector:
The module (amplitude) of \( {{\bf G}}\left[ {f\left( {x,y} \right)} \right] \) is:
The argument of \( {{\bf G}}\left[ {f\left( {x,y} \right)} \right] \) is:
From a perspective of edge detection, B x and B y denote the vertical and horizontal edge convolution kernel so-called B-spline gradient operator (BSGO) in this paper. In edge detection, the gradient ∇f at point (x, y) can be computed by the following expression:
We first derive the gradient image from the original tilt image by BSGO, compute the mean value of the gradient image (i.e. the mean gradient value) used as the gradient threshold value to binarize the gradient image. Therefore the results of edge detection acquired by BSGO represent the image contours, which are selected as the feature points to explore the initial values for registration in this paper. Therefore, the edges of reference image R and floating image F are detected by BSGO and then the binarization images B R and B F are acquired respectively. Here, B R and B F , essentially denote the feature points.
3.2 Computation of the centroid of the medical image
Definition 2 For a 2-D discrete function g(x, y), the moment of order (p + q) is defined as [10]
where parameter (p + q) is the order of the moment; M and N represent the numbers of sampling points in space.
Definition 3 The zeroth is expressed as [10]
All the first- and higher-order moments divided by M 0,0 is independent of the gray level of an object.
Definition 4 When p = 1 and q = 0, and, p = 0 and q = 1[10],
where (\( \overline x \) , \( \overline y \)) is defined as the centroid coordinates of the object.
When registering the images, according to Eqs. (17), (18) and (19), the zeroth and first-order moments of the image B R and image B F , are computed respectively, and then the centroids (\( \overline {{{x}_R}} \),\( \overline {{{y}_R}} \)) and (\( \overline {{{x}_F}} \),\( \overline {{{y}_F}} \)) are procured.
3.3 Acquisition of the rotation angle of the medical image
In the process of engineering design and experimental statistics, upon analysis and processing of a batch of data, the data are often fitted to a curve. LSM is a widely-used fitting method. If we use a smooth curve y = f(x) to fit a set of data \( \left( {{{x}_i},{{y}_i}} \right)\left( {i = 1,2, \cdots, n} \right) \), then in principle, the deviation between the data and the curve should be minimized. The deviation \( {{\varepsilon }_i} = f\left( {{{x}_i}} \right) - {{y}_i} \) is usually called residue. LSM enables the sum of \( \varepsilon_{{_i}}^2 \) to achieve the minimum. Here, we use a straight line to fit the data. Let the linear equation be expressed as \( y = {{a}_0}x + {{a}_1} \), the objective function Q:
The matrix form of Eq. (20) is shown as follows:
where \( {{\bf A}} = {{\left[ {\matrix{ {{{a}_0}} &{{{a}_1}} \\ }<!end array> } \right]}^T} \), \( {{\bf X}} = {{\left[ {\matrix{ {{{x}_1}} &{{{x}_2}} & \cdots &{{{x}_n}} \\ 1 & 1 & 1 & 1 \\ }<!end array> } \right]}^T} \), and \( {{\bf Y}} = {{\left[ {\matrix{ {{{y}_1}} &{{{y}_2}} & \cdots &{{{y}_n}} \\ }<!end array> } \right]}^T} \).
From Eq. (21), the partial derivative of A is obtained:
The above equation is solved and A is obtained:
Namely:
For a straight line, apparently, its tilt angle ϕ
In general, the method obtaining the rotation angle of the reference image R is delineated as follows:
-
Step 1
According to Eqs. (17), (18) and (19), the centroid (\( \overline {{{x}_R}} \),\( \overline {{{y}_R}} \)) of the binarization image B R is computed;
-
Step 2
The tilt matrix \( {{{{P}}}_{{{R}}}} \in {{R}^{{2 \times \left( {M \times N} \right)}}} \) is built, which denotes a 2-D matrix of coordinates (x,y) in the image B R :
$$ \left\{ {\matrix{ {{{P}_R}\left( {{1},\left( {i - {1}} \right) \times N + j} \right) = \left( {i - \overline {{{x}_R}} } \right) \times {{B}_R}\left( {i,j} \right)} \\ {{{P}_R}\left( {{2},\left( {i - {1}} \right) \times N + j} \right) = \left( {j - \overline {{{y}_R}} } \right) \times {{B}_R}\left( {i,j} \right)} \\ }<!end array> } \right. $$(26)where \( i = 1,2, \cdots, M;j = 1,2, \cdots, N \).
-
Step 3
Let n = M × N, put P R into Eq. (24) and α 0 is obtained;
-
Step 4
According to Eq. (25), the rotation angle ϕ R is derived.
Getting the rotation angle of floating image F has the same process as that of reference image R.
3.4 Similarity metric between two images
From the perspective of image restoration, medical image registration is to restore the floating image with geometric distortion to the reference image. Therefore, there must be a criterion to be used for objectively evaluating the quality of the restored image. Although the MI well reflects the information of the overlapping region between two images, it cannot directly and globally exemplify the gray difference between the images. At the same time, it has a heavily computational load and a time-consuming operation (See Table 1 in Reference [18]).
Since PSNR can represent the distinction between images, so far it is an extensively-used criterion that gauges the quality of a restored image. Generally speaking, the greater the value PSNR, the smaller the difference between images, the more similar two images, and the better the quality of the restored image; contrarily, with a smaller value, the difference is greater, two images are more different, and the quality is poorer. PSNR is defined as
Compared with MI, PSNR enjoys a better time advantage (See Table 2 in Reference [18]). Therefore, we use PSNR as the similarity metric between the reference and floating images in this paper. Herein Eq. (3) is rewritten
That is, searching an optimal transformation T optimal maximizes the PSNR of the registered floating image. In the actual image registration, generally, \( \sum\limits_{{x = 1}}^M {\sum\limits_{{y = 1}}^N {{{{\left( {r\left( {x,y} \right) - f\left( {x,y} \right)} \right)}}^2}} } = 0 \) is almost impossible in Eq. (27). However, theoretically, \( \sum\limits_{{x = 1}}^M {\sum\limits_{{y = 1}}^N {{{{\left( {r\left( {x,y} \right) - f\left( {x,y} \right)} \right)}}^2}} = 0} \) is possible in the process of the image self-registration. For instance, Fig. 1 is a reference image. We impose the self-registration on the image in Fig. 1 by applying the X, Y coordinate translations and image rotation, respectively, and the three change curves of PSNR are shown in Fig. 2.
As shown in Fig. 2, we can see clearly the breaks in the change curves of PSNR at the zero-crossing point of the horizontal axis, which results from \( \sum\limits_{{x = 1}}^M {\sum\limits_{{y = 1}}^N {{{{\left( {r\left( {x,y} \right) - f\left( {x,y} \right)} \right)}}^2}} = 0} \) and then \( PSNR = \infty \) in the process of the image self-registration. Equation (27), therefore, must be modified with a complementary value. Then we get the MPSNR according to Reference [18] as follows
According to Eq. (29), applying the image self-registration to Fig. 1 gets the change curves of MPSNR, shown in Fig. 3. According to the Fig. 3, due to the presence of the complementary value \( \frac{{{{{\left( {L - 1} \right)}}^2}}}{{10}} \), there exist no breaks in the change curves of MPSNR and each curve is smooth and continuous.
3.5 Procedure of medical image registration based on LSM and MPSNR
As mentioned before, medical image registration based on LSM and MPSNR (RLM) is detailedly depicted as follows:
-
Step 1
According to BSGO, the binarization images B R and image B F are acquired.
-
Step 2
According to Eqs. (17), (18) and (19), images B R and B F , the centroids (\( \overline {{{x}_R}} \),\( \overline {{{y}_R}} \)) and (\( \overline {{{x}_F}} \),\( \overline {{{y}_F}} \)) of reference image R and floating image F, are computed respectively.
-
Step 3
According to LSM, images B R and B F , the rotation angles ϕ R and ϕ F of reference image R and floating image F, are obtained respectively.
-
Step 4
The initial values for registering the images are calculated, namely, \( \Delta {{x}_0} = \overline {{{x}_F}} - \overline {{{x}_R}} \), \( \Delta {{y}_0} = \overline {{{y}_F}} - \overline {{{y}_R}} \), \( \Delta {{\varphi }_0} = {{\varphi }_{{{F}}}} - {{\varphi }_{{{R}}}} \).
-
Step 5
Choosing MPSNR as the similarity metric, and the simplex method as a multi-parameter optimizing method, is applied to get the translation and rotation parameters.
-
Step 6
The optimization process ends and the optimal transformation T optimal is procured.
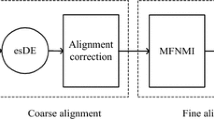
Now, we discuss the difference between RLM and RSMP in Reference [18]. They are two different approaches to obtaining the initial values for image registration. The proposed method in this paper uses BSGO to detect the edges of the original reference and floating images and then the binarization images are acquired. By computing the binarization image moments, the centroids are obtained. Also, according to the binarization image coordinates, the rotation angles of the reference and floating images are computed respectively, on the foundation of which the initial values for registering the images are produced. As for RSMP, it computes the original medical image moments to obtain the centroid, and according to the rotational invariance of the singular values of the matrix of the medical image coordinates, the rotation angles of the reference and floating images are computed respectively, on the foundation of which the initial values for registering the images are generated.
4 Experiments and results
In this paper, all the experimental images are extracted from the brain image database built by Retrospective Registration Evaluation Projection, which is affiliated to Vanderbilt University, USA. RLM and MIR are implemented in MATLAB 7.1 on PC with a Pentium IV 2.6 GHz and 1,024 MB RAM, running Windows XP. In order to validate the features that RLM has a fast implementation, a high registration precision and a strong robustness, we use the simplex method as multi-parameter optimizing one, and compare the results from the MIR.
In the following sections, we choose No.25 CT, No.24 MR_T1_rectified and No.10 PET brain images of the patient_001 as the reference images, whose sizes are 512 × 512, 256 × 256 and 128 × 128, respectively, and gray levels are all represented by 256, shown in Fig. 4a, b, and c.
To compare the accuracy of the image registration, we define error ρ i [18]
where ∆i s denotes the standard transformation parameter aligning the floating image with the reference image, and ∆i represents the actual transformation parameter acquired by the registration method mentioned in this paper. Further, we define total error ρ [18]
4.1 Mono-modality medical image registration
In the section, we choose No.25 CT, No.24 MR_T1 and No.10 PET brain images of the patient_001 as the reference images, shown in Fig. 4a, b, and c, respectively. Each reference image is transformed into the corresponding floating image according to the parameters in Table 1, shown in Fig. 5a and b, Fig. 6a and b, and, Fig. 7a and b. In the following registered images, the red and green landmarks denote the results from extracting the edges of the reference and floating images by Canny operator respectively, and the yellow landmarks denote the overlapping region of two registered images.
By Applying image moments and LSM ∆x 0, ∆y 0 and ϕ 0 are excavated to be used for the initial values of the simplex method shown in Table 2, and then RLM and MIR are performed. In order to compare the proposed method, ∆x 0, ∆y 0 and ϕ 0 are obtained by image moments and the singular value decomposition (SVD) in Reference [18] shown in Table 3, and then RSMP is implemented. The experimental results are listed in Figs. 8, 9 and 10 and Table 4.
From Table 2, the initial values derived from images moments and LSM are approximate to the transformation parameters in Table 1 and the time cost is also very low, but the accuracies need to be further promoted, which lay on a solid foundation for image registration. In addition, it should be noted at this point that, the initial values in Table 3 generated by images moments and SVD are superior to those by images moments and LSM, the calculation load is much lower, and the accuracies are also higher. Therefore, images moments and SVD can provide the more outstanding initial values in the mono-modality medical image registration.
According to the data in Table 4, the registration speed of RLM is obviously superior to that of the MIR, especially for the larger image, but is inferior but closer to that of RSMP. According to Eqs. (30) and (31), we also compute the registration accuracies of RLM, MIR and RSMP, and the errors are shown in Table 4. The results reveal that, the registration accuracy of RLM, on the whole, achieves that of MIR or even better. As compared with RSMP, the accuracy of RLM has no advantage, but their discrepancy is tinier. As stated above, selecting MPSNR as the similarity metric is a better and more effective strategy. In addition, we also can find, since the initial values generated by image moments and LSM are close to the standard transformation parameters in Table 1, RLM and MIR can quickly and exactly obtain the final transformation parameters. Therefore, as images moments and SVD, images moments and LSM used for acquiring initial values before registration is also a good preprocess.
4.2 Multi-modality medical image registration
In the section, we still use No.25 CT, No.24 MR_T1 and No.10 PET brain images of the patient_001 as the reference images. The experimental images are divided into the following three groups. The first is to select CT image as the reference one and, MR1 and MR2 as the floating images, whose sizes are of 256 × 256 pixels, shown in Fig. 11. The second is to select CT image as the reference one and, PET1 and PET2 as the floating images, whose sizes are of 128 × 128 pixels, shown in Fig. 12. And the last is to select MR image as the reference one and, PET1 and PET2 as the floating images, whose sizes are of 128 × 128 pixels, shown in Fig. 13. In the experiments, RLM, MIR and RSMP are applied to register the multi-modality images respectively, and we still use the simplex method to search the optimal registering parameters. Also, we list the relatively accurate transformation parameters in Table 5, which are taken as ∆i s in Eq. (30).
∆x 0, ∆y 0 and ϕ 0 acquired by applying image moments and LSM, are still used for the initial values of the simplex method, shown in Table 6. In order to compare the proposed method, ∆x 0, ∆y 0 and ϕ 0 are obtained by image moments and SVD in Reference [18] shown in Table 7, and then RSMP is implemented. The experimental results based on RLM, MIR and RSMP are shown in Figs. 14, 15 and 16 and Table 8, respectively.
From Tables 6 and 7 we can know, the use of image moments and LSM is still an efficient strategy acquiring the initial values for registration, and its calculation load is inferior to that of RSMP. Surprisingly, on the whole, in the multi-modality medical image registration, image moments and LSM can provide the more outstanding initial values than images moments and SVD.
According to the registering results from Table 8, the registering speed of RLM has is still considerably superior to that of MIR, especially for the larger image, and is inferior but closer to that of RSMP. According to Eqs. (30) and (31), we obtain the registration errors of RLM, MIR and RSMP, listed in Table 8. The results indicate that, the registration accuracy of RLM, overall, achieves that of MIR or even better. As compared with RSMP, the accuracy of RLM has some advantage. Therefore, selecting MPSNR as the similarity metric in the multi-modality image registration is the same effective criterion as MI. In addition, the method getting the initial values is devoted to RLM and MIR, and reduces a considerable amount of time.
5 Discussions
As illustrated above, RLM has an excellent registration performance, which is derived from both getting the initial values for registration by image moments and LSM, and selecting the MPSNR as the similarity metric. The former can advance the performance not only of the intensity-based but also of the feature-based registration methods according to the experimental results. Among the feature-based registration methods, the ICP algorithm pioneered by Besl and Mckay [2] in 1992, is one well known approach and extensively used for the registration on point sets. It can register the point sets without any prior and explicit segmentation or other preprocessing of object features, which makes it to be prevalent and very well adapted for point set registration. In the past twenty years, the ICP algorithm has made significant progress, and has further been complemented and perfected. Chen, Medioni [7] and Bergevin et al. [1] proposed a precise registration method that finds the closest points based on point-to-plane distances. Rusinkiewicz and Levoy [24] introduced a fast registration method that searches the closet points using point-to-projection metric. Park and Subbarao [19] developed a registration approach that explores the closest points based on contractive-projection-point metric. In addition, Gelfand et al. [9] analyzed the quality of registration for point cloud data.
Although ICP algorithm is widely used for image registration, it has following three key problems: 1) It has heavily computational load in the process of searching the corresponding closest points; 2) Whether it successfully gets the optimal registration parameters strongly depends on the initial rotation and translation matrixes selected. When they are ill-suited, the registration process needs more searching time, easily gets into the local optima, and even fails to register images; And 3) it is very difficult to automatically acquire the crucial feature points in image registration. In this paper, the ICP algorithm incorporating with the method acquiring the initial values, an improved iterative closest point algorithm based on acquiring the initial values for registration from the least square method (LICP) is introduced. LICP first utilizes BSGO to detect image edges, and then produces a binarization image to automatically acquire the feature points, finally uses image moments and LSM to obtain the centroids and rotation angles of the reference and floating images respectively. As stated above, LICP is delineated in detail as follows:
-
Step 1
According to BSGO, the binarization images B R and image B F are acquired.
-
Step 2
According to Eqs. (17), (18) and (19), images B R and B F , the centroids (\( \overline {{{x}_R}} \),\( \overline {{{y}_R}} \)) and (\( \overline {{{x}_F}} \),\( \overline {{{y}_F}} \)) of the reference image R and the floating image F, are computed respectively.
-
Step 3
According to LSM, images B R and B F , the rotation angles ϕ R and ϕ F of the reference image R and floating image F, are obtained respectively.
-
Step 4
The initial values for registering the images are calculated, namely, \( \Delta {{x}_0} = \overline {{{x}_F}} - \overline {{{x}_R}} \), \( \Delta {{y}_0} = \overline {{{y}_F}} - \overline {{{y}_R}} \), \( \Delta {{\varphi }_0} = {{\varphi }_{{{F}}}} - {{\varphi }_{{{R}}}} \).
-
Step 5
Δx 0, Δy 0 and Δϕ 0 are used as the initial translation and rotation parameters for the ICP algorithm, namely
$$ {{{{\bf R}}}_0} = \left[ {\matrix{ {\cos \left( {\Delta {{\varphi }_0}} \right)} &{ - \sin \left( {\Delta {{\varphi }_0}} \right)} \\ {\sin \left( {\Delta {{\varphi }_0}} \right)} &{\cos \left( {\Delta {{\varphi }_0}} \right)} \\ }<!end array> } \right],{{{{\bf T}}}_0} = {{\left[ {\matrix{ {\Delta {{x}_0}} &{\Delta {{y}_0}} \\ }<!end array> } \right]}^T} $$ -
Step 6
Two sets of coordinates representing all the pixels with gray values being 1 or 255 in the images B R and B F respectively, are obtained, and they are selected as the reference and floating point sets in the ICP algorithm.
-
Step 7
The ICP algorithm is performed and the final rotation matrix R 0 and translation T 0 are derived.
5.1 Mono-modality medical image registration
In the section, we still use the experimental images in Section 4.1. The ICP algorithm, LICP and IICP [18] are applied to register the mono-modality medical images, respectively, and the experimental results are shown in Figs. 17, 18 and 19 and Table 9.
From Table 9, the registration speed of LICP is obviously superior to that of the ICP, especially for the larger image, but is far inferior to that of IICP. From Figs. 17, 18 and 19, the ICP algorithm fails to register the images, while LICP and IICP register all the images successfully, and the registration accuracy of IICP is superior to that of LICP in the mono-modality registration, which are in accordance with the errors shown in Table 9. As seen above, the use of acquiring initial translation and rotation parameters for the ICP algorithm is also a good and effective approach.
5.2 Multi-modality medical image registration
In the section, we still use the experimental images in Section 4.2. The ICP algorithm, LICP and IICP are applied to register the multi-modality medical images, respectively, and the experimental results are shown in Figs. 20, 21 and 22 and Table 10.
From Table 10, the registration speeds of LICP and IICP are relatively superior to that of the ICP, especially for the larger image. From Figs. 20, 21 and 22, the ICP algorithm fails to register the images; IICP fails to align PET1 with CT and PET2 with CT, while LICP is successful in registering all the images, which are in accordance with the errors in Table 10. However, we also can find, the accuracy of IICP is inferior to that of LICP in the multi-modality registration. As stated above, the use of the method generating initial values is still an effective measure.
5.3 Comparisons of RLM and LICP
RLM and LICP, as explained above, due to the use of obtaining the initial values for registration by image moments and LSM, their registration accuracies are greatly advanced and processing times are significantly reduced. In addition, RLM also benefits from the new, fast similarity metric, and LICP also takes advantage of the automatic feature point extraction by BSGO. However, RLM and LICP are still obvious differences in the performance.
In the mono-modality image registration, both RLM and LICP can successfully register all the images. With respect to the registration accuracy, RLM outperforms LICP except in the case of aligning MR2 with MR. Considering the processing time, LICP is superior to RLM in addition to the alignment of MR1 with MR. Especially for registering larger images, the difference between the processing times is more considerable. In the multi-modality image registration, RLM and LICP can successfully register all the images. As for the registration accuracy, RLM is a more excellent method compared with LICP, while LICP has a relatively greater errors and its accuracy needs to be further increased. Concerning the processing time, LICP is still superior to RLM.
6 Conclusions
In order to solve the existed problems of registering the images based on MI, by using image moments and LSM as the tool for getting the initial values for registration, MPSNR as the similarity metric between the reference and floating images, and the simplex method as the optimizing means, medical image registration based on LSM and MPSNR is introduced. The experimental results reveal that, RLM has a simple logical structure, a short computation, a fast registration and a high accuracy, and also overcomes the problem of easily getting into the local optimum. It can be well suitable for both mono-modality and multi-modality image registrations. In addition, the use of obtaining the initial values also can help the ICP algorithm to enhance the performance. The research of employing RLM to the non-rigid medical image registration will be our next focus.
References
Bergevin R, Soucy M, Gagnon H et al (1996) Towards a general multi-view registration technique. IEEE Trans Pattern Anal Mach Intell 18(5):540–547
Besl PJ, Mckay ND (1992) A method for registration of 3-D shapes. IEEE Trans Pattern Anal Mach Intell 14(2):239–256
Blondel C, Vaillant R, Malandain G et al (2004) 3D tomographic reconstruction of coronary arteries using a precomputed 4D motion field. Phys Med Biol 49(11):2197–2208
Borgefors G (1988) Hierarchical chamfer matching: a parametric edge matching algorithm. IEEE Trans Pattern Anal Mach Intell 10(6):849–865
Brown L (1992) A survey of image registration techniques. ACM Comput Surv 24(4):325–376
Can A, Stewart C (2002) A feature-based, robust, hierarchical algorithm for registration palm of images of the curved human retina. IEEE Trans Pattern Anal Mach Intell 24(3):347–363
Chen Y, Medioni G (1992) Object Modeling by Registration of Multiple Range Images. Imag Vis Comput 10(3):145–155
Fei B, Wheaton A, Lee Z et al (2002) Automatic MR volume registration and its evaluation for the pelvis and prostate. Phys Med Biol 47(5):823–838
Gelfand N, Ikemoto L, Rusinkiewicz S, et al. (2003) Geometrically Stable Sampling for the ICP Algorithm. Proceedings of the 4th International Conference on 3-D Digital Imaging and Modeling. Stanford University, CA, USA, 260–267
Hu M (1962) Visual pattern recognition by moment invariants. IRE Trans Inform Theory 8:179–187
Hyunjin P, Peyton H, Kristy K et al (2004) Adaptive registration using local information measures. Med Image Anal 8(4):465–473
Krucker JF, LeCarpentier GL, Fowlkes JB et al (2002) Rapid elastic image registration for 3-D ultrasound. IEEE Trans Med Imag 21(11):1384–1394
Kybic J, Unser M (2003) Fast parametric elastic image registration. IEEE Trans Image Process 12(11):1427–1442
Lehmann TM, Gonner C, Spitzer K (1999) Survey: Interpolation Methods in Medical Image Processing. IEEE Trans Med Imaging 18(11):1049–1075
Lu ZT, Feng YQ, Feng QJ et al (2008) Medical Image Registration Based on Principal Phase Congruency. Acta Electronica Sinica 36(10):1974–1978
Maes F, Collignon A, Vandermeulen D et al (1997) Multimodality image registration by maximization of mutual information. IEEE Trans Med Imag 16(2):187–198
Maintz JBA, Viergever MA (1998) A survey of medical image registration. Med Image Anal 2(1):1–36
Pan MS, Tang JT, Yang XL (2011) Medical Image Registration Based on SVD and Modified PSNR. J Inf Sci Eng 27(5):1761–1775
Park SY, Subbarao M (2004) Automatic 3D model reconstruction based on novel pose estimation and integration techniques. Imag Vis Comput 22(8):623–635
Paul V, William M, Wells I (1997) Alignment by Maximization of Mutual Information. Int J Comput Vis 24(2):137–154
Pluim JPW, Maintz JBA, Viergever MA (2003) Mutual-information-based registration of medical Images: A Survey. IEEE Trans Med Imaging 22(8):986–1004
Radau PE, Slomka PJ, Julin P et al (2001) Evaluation of linear registration algorithm for brain SPECT and the errors due to hypoperfusion lesions. Med Phys 28(8):1660–1668
Rueckert D, Sonoda L, Hayes C et al (1999) Nonrigid registration using free-form deformations:application to breast MR images. IEEE Trans Med Imag 18(8):712–721
Rusinkiewicz S, Levoy M (2001) Efficient variants of the ICP algorithm. Proceedings of the 3rd International Conference on 3-D Digital Imaging and Modeling, Quebec, 145–152
Saber E, Tekalp AM (1997) Region-Based Shape Matching for Automatic Image Annotation and Query-by-Example. J Vis Comm Image Represent 8(1):3–20
Slomka PJ, Mandel J, Downey D et al (2001) Evaluation of voxel-based registration of 3-D power doppler ultrasound and 3-D magnetic resonance angiographic images of carotid arteries. Ultrasound Med Biol 27(7):945–955
Stammberger T, Eckstein F, Michaelis M et al (1999) Interobserver reproducibility of quantitative cartilage measurements: comparison of B-spline snakes and manual segmentation. Magn Reson Imag 17(7):1033–1042
Staring M, Klein S, Pluim JPW (2007) Nonrigid registration with tissue-dependent filtering of the deformation field. Phys Med Biol 52(23):6879–6892
Szeliski R, Coughlan J (1997) Spline-based image registration. Int J Comput Vis 22(3):199–218
Thevenaz P, Unser M (2000) Optimization of mutual information for multiresolution image registration. IEEE Trans Image Process 9(12):2083–2099
Unser M (1999) Spline: A perfect fit for signal and image processing. IEEE Signal Process 16(8):22–38
Unser M, Aldroubi A, Eden M (1993) B-spline Signal Processing: Part-I Theory. IEEE Trans Signal Process 41(2):821–833
Unser M, Aldroubi A, REden M (1993) B-spline Signal Processing: Part-II Theory. IEEE Trans Signal Process 41(2):834–848
Acknowledgement
This paper was partially supported by the Key Discipline-Leader Foundation of Hunan Provincial Institutions of Higher Education, PRC and supported by Outstanding Young Scientific Research Fund of Hunan Provincial Education Department, PRC(No.09B071).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pan, Ms., Jiang, Jj., Rong, Qs. et al. A modified medical image registration. Multimed Tools Appl 70, 1585–1615 (2014). https://doi.org/10.1007/s11042-012-1180-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-012-1180-3