Abstract
Background
Shell color formation is an important physiological process in bivalves, the molecular genetic basis has potential application in bivalve aquaculture, but there is still remaining unclear about this issue. The cystine/glutamate transporter (Slc7a11) and cystathionine beta-synthase (Cbs) are integral genes in pheomelanin synthesis pathway, which is vital to skin pigmentation.
Methods and results
Here, the sequences of b (0, +) -type amino acid transporter 1 (B-aat1) and Cbs in Pacific oyster (Crassostrea gigas) (CgB-aat1, CgCbs) were characterized. Phylogenetically, the deduced amino acid sequences of CgB-aat1 and CgCbs both possessed conserved features. Genes were both ubiquitously expressed in six tested tissues with more abundant expression level in central mantle. Besides, the polyclonal antibodies of CgB-aat1, CgCbs, CgTyr, and CgTyrp2 were successfully prepared. Immunofluorescence analysis revealed that CgB-aat1 and CgCbs proteins were both expressed in gill rudiments of eyed-larvae and concentrated mainly in cytoplasm of epithelial cell and nerve axons in mantle. Additionally, after CgB-aat1 or CgCbs silencing, expressions at mRNA and protein levels of CgB-aat1 and CgCbs involved in pheomelanin synthesis were significantly suppressed, and CgTyr, CgTyrp1 and CgTyrp2 related to eumelanin synthesis were also down-regulated but no apparent differences, respectively. Moreover, micrographic examination found less brown-granules at mantle edge in CgB-aat1 interference group.
Conclusion
These results implied that pheomelanin synthesis was possible induced by CgB-aat1-CgTyr-CgCbs axis, and it played an essential role on mantle pigmentation in the oysters. These findings provide the useful genetic knowledge and enrich the physiological information for the shell color formation in bivalve aquaculture.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The colorful molluscan shells are widely recognized and have been appreciated by collectors and scientists. Colorful shell is mainly because of the biological pigments. Melanin, one of the most common pigments, include black eumelanin (brown/black pigment) and lighter pheomelanin (orange/red pigment). Both of them was found in mammals, birds and insects, and only eumelanin was found in fish [1]. However, melanin in molluscan shells has not been fully characterized due to the fact that it is difficult to isolate [2]. Until now, Affenzeller et al. [3] found the existence of pheomelanin in the terrestrial gastropod Cepaea nemoralis using HPLC.
In addition, the biosynthetic and molecular pathway of melanin synthesis are well not understood in mollusks, except in insects and mammals [1, 4]. Generally, Tyr initiates eumelanogenesis by hydroxylating tyrosine to dopa and oxidizing the resultant dopa to dopaquinone, which is then converted to eumelanin polymer by Tyrp1 and Tyrp2 [5]. On the other hand, SLC7a11/xCT can transport cystine into cell. Subsequently, cystine and dopaquinone are catalyzed by CBS and consequently promote the synthesis of pheomelanin [1, 6]. Recently, omics analysis in different color varieties of fishes have been done to confirm the conserved melanogenesis pathway in fish [7,8,9]. Furthermore, the sequences, expressions and functions of Slc7a11/xCT involved in skin color have been characterized in vertebrate [10,11,12,13]. And the structural characterize of Cbs is also well studied that has crucial role in transsulfuration for cysteine biosynthesis [14]. These works demonstrated that Slc7a11/xCT and Cbs act vital roles in pheomelanin synthesis pathway. To date, the complete pathway of melanin synthesis in mollusks is not known, several genes such as Tyr, Tyrp2, Mitf and Pax3/7 have been shown to be important in the regulation and production of melanin in mollusks [15,16,17]. However, other research on molecular pathway of pheomelanin synthesis in mollusks are still poorly understood and no study to date has addressed these issues.
The Pacific oyster, Crassostrea gigas, is one of the most economically important marine bivalve species. Recently, oysters with orange shell both of the right and left, a recessive trait compared to black and white, were obtained from breeding program [18]. The whole transcriptome analysis revealed that melanin biosynthesis-related genes functioned on orange shell coloration in C. gigas [19]. Currently, existing research was limited to quantification of melanin synthesis related genes in C. gigas. Hence, it is urgent to study the molecular characteristics and function in shell color of these genes.
Here, B-aat1 and Cbs gene from orange shell color oysters were characterized and expressed. Furthermore, gene function was deliberated by RNAi technology, one of the methods of transporting dsRNA was bacterial feeding, which has been well applied in C. gigas [20, 21]. These results will help understand the overall role of B-aat1 and Cbs in pheomelanin synthesis pathway and mantle pigmentation as well as advance our knowledge of shell color genetics in bivalve.
Materials and methods
Oysters collection
The Pacific oyster is neither an endangered nor protected species. All experiments in this study were conducted according to national and institutional guidelines. Here, one-year old oysters with orange shell color were supplied with filtered seawater at ambient temperature (24 ± 1 °C), salinity (30.5 ± 0.5 psu) and pH (8.1), which was the same with condition in farm of Rongcheng, Shandong province, China. Seven days later, the oysters (shell length 24.02 ± 8.09 cm and shell height 36.55 ± 9.08 cm) were chosen for sampling. Six tissues, including digestive gland, labial palp, gill, mantle edge, central mantle, and adductor muscle were dissected and immediately stored at − 80 °C for RNA extraction. In embryonic and larval sampling, oysters with orange shell color were transported to Litao hatchery, Yantai City, Shandong Province. The embryos were obtained following our previous study [21]. In brief, after mixing sperm and oocytes, fertilized eggs were reared with temperature (23–25 °C) and salinity (30–31 psu). Isochrysis galbana and Chaetoceros calcitrans were mixed to feed larvae daily. In a month, fertilized egg, blastula, gastrula, trochophore, D-shaped larvae 1/2, umbo larva 1/2 and eyed-larva were collected for RNA extractions and IF analysis.
Bioinformatics analysis
The amino acid (aa) sequences of b (0, +) -type amino acid transporter 1 (B-aat1) and cystathionine beta-synthase (Cbs) in C. gigas (CgB-aat1, XP_034337681.1; CgCbs, XP_034300359.1) and B-aat1/Slc7a11 as well as Cbs from other organisms were retrieved from NCBI database (Table 1). The SMART database (http://smart.embl-heidelberg.de/) was used to predict gene function domain. The TMHMM-2.0 database (https://services.healthtech.dtu.dk/service.dtu.dk/service.php? TMHMM-2.0) was used to analysis transmembrane helices of CgB-aat1 protein. Multiple sequence alignment of CgB-aat1 and CgCbs proteins with other known B-aat1/Slc7a11 and Cbs proteins was performed using DNAMAN 8.0 software. The alignment of the sequences was analyzed using Clustal X 2 and phylogenetic tree was constructed with the method of maximum-likelihood algorithm using MEGA X.
Gene expression analysis
The cDNA template used for qPCR was synthesized using PrimeScript™ RT reagent Kit (Takara, China). The Primer3 from NCBI (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) was used to design qPCR primer pairs (listed in Table S1), in which ef1α gene was set as reference gene [22]. The qPCR reactions were performed with QuantiNovaTM SYBR® Green PCR Kit (QIAGEN, Germany) on LightCycler 480 real-time PCR instrument (Roche, Switzerland). Three repeats of each cDNA sample were reacted in 10 μL volume, respectively, which contained 5 μL 2× SYBR Green PCR Master Mix, 1 μL cDNA template, 2 μL ddH2O and 1 μL each 10 mM primer. The PCR reaction were as follows: 95 °C for 2 min, followed by 40 cycles of 95 °C for 5 s and 60 °C for 10 s. All primers specificity were confirmed by melt cure (65–95 °C) analysis. The 2–ΔΔCT method was applied to gene expression analysis [23]. All data were shown as means ± standard error (SE) (n = 6). Statistically significant differences were analyzed using t-test and one-way ANOVA method between two data and among multiple data, respectively. The significance level was considered at P < 0.05.
Antibody preparation and WB examination
The recombinant CgCbs, CgTyr and CgTyrp2 protein was generally conducted respectively as described by Liu et al. [24]. For CgB-aat1, we generated the antibody again rabbit polyclonal antibody from a 11-residue polypeptide (CGEPQIPKKMI). The other purified CgCbs/ CgTyr/ CgTyrp2 recombinant protein was used to immunize two rabbits. In brief, four immunizations were performed at 8 days intervals. After immunization twice, antiserum collection and ELISA assay was conducted weekly to assess the immunity level. Following the titer evaluation, antibody specificity was verified by WB as described below.
Purified proteins and antibody obtained were detected using WB. In details, recombinant proteins (20 μg) were separated on 12.5% (v/v) polyacrylamide vertical slab gel and transferred onto polyvinyl difluoride membrane (Beyotime, Shanghai, China). Following blocking with 5% skimmed milk at 25 °C for 2 h, the membrane was incubated with His-flag (Beyotime, China) /CgB-aat1/CgCbs/CgTyr/CgTyrp2 polyclonal antibody (diluted 1:2000 in Blocking Solution) at 4 °C overnight and then incubated with HRP-conjugated goat anti-rabbit IgG (Beyotime, China) at 37 °C for 2 h. The chemiluminescence reaction was performed using the enhanced ECL chemiluminescence reaction detection kit (Vazyme, China) and protein bands were imaged with Gel Image System (JS-2000).
Immunofluorescence
IF examinations of CgB-aat1 and CgCbs proteins in eyed-larva and mantle tissue were performed according to the description by Wang et al. [10] with some modifications. Simply, the mantle was treated with fixation in Bouin reagent, dehydration in ethanol, transparent in xylol, embedding in paraffin and section in 5 μm thickness. Then, the slices were incubated in 0.01 M citrate buffer (pH = 6) at 100 °C for 10 min for antigen retrieval. After blocking with 7.5% goat serum (Beyotime), the primary antibody (anti-CgB-aat1 antibody with 1:1000 dilution and anti-CgCbs antibody with 1:500 dilution) incubation was performed at 4 ℃ overnight, then the samples were hatched with Alexa fluor® 488-labbled Goat Anti-Rabbit secondary antibody (1:500 dilution, cat. nos. A0423, Beyotime) at room temperature for 90 min. The normal goat serum incubation was set as the negative control. Cell nuclei were counterstained with DAPI (Beyotime). Finally, samples were captured using confocal microscopy (TCS SP98, Leica, Germany).
Synthesis and feeding of dsRNA
The fragments of CgB-aat1 and CgCbs genes were produced by PCR with primers listed in Table S1. Sub-cloning, ligation, transfection and induced expression were used for dsRNA synthesis base on the detailed description by Feng et al. [20]. Here, after transformation with three constructed plasmids (CgB-aat1-L4440, CgCbs-L4440 and EGFP-L4440), respectively, bacteria (Escherichia coli strain HT115) were induced by IPTG for producing three dsRNAs, respectively. EGFP was used as a negative control (NC). The non-induced E. coli and IPTG-induced E. coli transformed were collected for RNA isolation using Bacteria RNA Extraction Kit (Vazyme, China), respectively.
One-year-old oysters with orange shell were used for dsRNA feeding experiment after acclimating for 10 days at the same environment mentioned in Part 2.1. In which, oysters with shell length of 23.67 ± 8.03 cm and shell height of 33.75 ± 7.64 cm were selected. In this study, ten oysters in CgB-aat1-RNAi group, CgCbs-RNAi group and EGFP-RNAi group were carried out, respectively. During the feeding period, bacteria (E. coli strain HT115) containing recombinant plasmid were cultivated in LB medium at 37 °C overnight. The bacteria were diluted 100-fold in 500 mL medium and cultured until the OD595 values reached 0.4. Subsequently, 0.4 mM IPTG was added and incubated for 4 h to induce dsRNA production. Bacterial pellets obtained were re-suspended in 500 mL Platymonas subcordiformis and 125 mL Nitzschia closterium f. minutissima algae culture liquid. Then, oysters were fed with 625 mL Alga/dsRNA-producing bacteria co-inoculum once a day and fed with only alga twice a day. The 60% of seawater volume was changed daily with aerated seawater (24 ± 1 °C). After 40 days feeding, the dead oyster was not observed and mantles collected were used for RNA extraction, gene and protein expressions and histology examination.
Protein expression in mantle of CgB-aat1-RNAi group, CgCbs-RNAi group and EGFP-RNAi group (denoted as B-aat1-i, Cbs-i and Egfp-i, respectively) were analyzed using WB. In detail, the mantles were homogenized in 1 × PBS buffer and centrifuged at 13,000 g for 15 min, then the supernatants collected was used to measure the protein concentration using the Nanodrop 2000 (Thermo scientific, USA) [25]. Protein with 20 μg was prepared for loading, which was carried out as described above. The GAPDH rabbit monoclonal antibody diluted 1000-fold was served as control. CgB-aat1, CgCbs, CgTyr, Tyrp1 (abcam ID: EPR21956) and CgTyrp2 polyclonal antibody were used to detected the corresponding protein expression level in three groups, respectively.
Histology analysis
Mantle tissues from oysters in three groups were sampled after RNAi experiment and were used for analyzing histological structure changes of mantle. The slices of mantle were obtained following the method mentioned in IF. Then the sections were stained with H&E and examined under an Olympus BX53 light microscope (Olympus Corporation, Japan).
Results
Identifications of CgB-aat1 and CgCbs protein
In the deduced 497 aa CgB-aat1 protein, ten transmembrane regions were predicted using TMHMM-2.0 database (Fig. S1). The deduced aa sequences of CgB-aat1 and CgCbs, both possess conserved features among multiple species (Fig. S2a, c). Phylogenetic analysis showed that CgB-aat1 clusters together with sequences from other mollusks, finally with vertebrates (Fig. S2b). Phylogenetic analysis of CgCbs revealed all Cbs genes were classified into two clusters, Cbs genes in vertebrates and invertebrates were classified to a big clade, respectively (Fig. S2d).
Expressions of CgB-aat1 and CgCbs
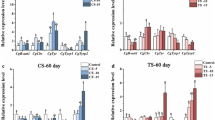
The expression of CgB-aat1 gene have distinct expression pattern in embryo-larval stages, significant increase of CgB-aat1 gene expression was detected at D-shaped 1 stage, where the expression upped to the top (P < 0.05) (Fig. 1a). The mRNA level of CgB-aat1 gene was widely distributed in six tissues, in which the highest expression was detected in central mantle, the lowest was in digestive gland (Fig. 1b). The results showed that CgCbs gene was also mainly expressed in eyed-larva (P < 0.05), with lower expression in the period before the blastula stage (P < 0.05) (Fig. 1c). CgCbs gene was also ubiquitously expressed in adult six tissues investigated with more abundant expression level in central mantle (Fig. 1d).
Expression analysis of CgB-aat1 and CgCbs genes estimated by qPCR. a, c Gene expression profiles of CgB-aat1 (a) and CgCbs (c) in six tissues. The digestive gland tissue was set as control. b, d Gene expression profiles of CgB-aat1 and CgCbs during embryonic and larval development. The fertilized egg was set as control. The significant difference (P < 0.05) was shown with different letters and all data were shown as mean ± standard error (SE) (n = 6)
Prokaryotic expression and antibody preparation
The expression of CgCbs/CgTyr/CgTyrp2 recombinant protein was successfully induced by IPTG in E. coli BL21 (DE3), SDS-PAGE result indicated that the target band migrated at about 18 kDa, 25 kDa, 20 kDa, respectively (line 3 in Fig. 2a, b, c, respectively). Purified recombinant CgCbs, CgTyr and CgTyrp2 proteins revealed a single band, respectively, and located at predicted molecular mass range (Fig. S3a–c). Purified protein was confirmed by WB, the result showed that purified protein with N-terminal His tag was specific to Rabbit anti-His-tag antibody (Fig. S3d). Polyclonal CgCbs/CgTyr/CgTyrp2 antibody was produced by immunizing rabbit with recombinant CgCbs/CgTyr/CgTyrp2 protein and the binding relationship with antigen was monitored by WB. After the specific antibody against the Cbs/Tyr/Tyrp2 fusion protein, a single band at around 18 kDa (Fig. 2d) / 25 kDa (Fig. 2e) / 20 kDa (Fig. 2f) was detected, respectively. Additionally, the specificity of Tyrp1 monoclonal antibody in mantle of C. gigas was confirmed by IF pretest (data not shown).
Production of recombinant CgCbs, CgTyr and CgTyrp2 proteins as well as WB validation of the antibodies. Determination of expression and purification of recombinant CgCbs (a), CgTyr (b) and CgTyrp2 (c) protein by SDS-PAGE analysis. M, molecular marker; Lane 1, BL21 without recombinant plasmid; Lane 2, protein expressed without IPTG induction (used as a negative control); Lane 3, protein expressed with IPTG induction for 6 h; Lane 4, soluble protein of supernatant; Lane 5, inclusion body; Lane 6, purified recombinant protein. The specificity of the CgCbs (d), CgTyr (e) and CgTyrp2 (f) antibody was verified by WB, respectively
Localization of CgB-aat1 and CgCbs proteins
IF analysis in eyed-larvae showed that CgB-aat1 protein mainly located in mantle and gill rudiments [Fig. 3a (b1–b2)]. And positive signals of anti-Cbs were observed in gill rudiments and anterior adductor muscle [Fig. 3a (c1)]. IF analysis results in mantle showed green fluorescence was mainly distributed on the cytoplasm of epithelial cell and nerve axons, indicating that CgB-aat1 and CgCbs proteins are mainly located on the cytoplasm in this organ (Fig. 3b). There was no signal was found in control groups conducted in both of eyed-larvae [Fig. 3a (d)] and mantle tissues [Fig. 3b (b1–b3)].
IF localization of CgB-aat1 and CgCbs proteins in eyed-larva (a) and in the mantle (b). a IF of competent eyed-larvae sections for CgB-aat1 and CgCbs protein (green fluorescence). The blue fluorescence is nucleus (N). Each fluorescent IF section is accompanied with section stained with H&E technique (a1–a2). aam: anterior adductor muscle; ao: apical organ; f: foot; gr: gill rudiments; m: mantle; pam: posterior adductor muscle; pg: pedal ganglion v: vellum with cilia; vc: visceral cavity; vm: vellum membrane. b1–c1 Sagittal section of whole competent larvae. Scale bar = 100 μm. b1–b2 Obvious positive signals of anti-CgB-aat1 were observed in mantle (m shown with white arrow in b1), gill rudiments (gr shown with white arrows in b2). The false positive was shown in the around of the visceral cavity. c1 Sagittal section of whole competent larvae. Stronger positive signals of anti-CgCbs were shown in gill rudiments and anterior adductor muscle, showing with white arrows. d1 Immunofluorescence localization analyses in negative control. The negative controls (NC) were incubated with normal goat serum. The white arrows mean false positive. b Positive signals of anti-CgB-aat1 and anti-CgCbs immunolabeling were shown in green fluorescence (a1, c1), which was mainly distributed on the cytoplasm of epithelial cell and nerve axons in mantle. The blue fluorescence is nucleus (N) (a2, c2). The negative controls (NC) were incubated with normal goat serum and there was no obvious positive signal observed in NC group. Scale bar = 100 μm. (Color figure online)
Expressions of CgB-aat1 and CgCbs were inhibited after RNA interference
Gene expressions levels of CgB-aat1 and CgCbs genes were reduced by 97.52% in CgB-aat1 dsRNA group (Fig. 4a) and 63.46% in CgCbs dsRNA group (P < 0.05) in comparison with EGFP dsRNA group (negative control, NC), respectively (Fig. 4b). WB results displayed that CgB-aat1/CgCbs protein was obvious depressed when compared with NC group (Fig. 4c). There was no doubt that CgB-aat1/CgCbs dsRNA feeding could effectively inhibit CgB-aat1/CgCbs expression.
Expressions of CgB-aat1, CgCbs, CgTyr, CgTyrp1 and CgTyrp2 after CgB-aat1 and CgCbs silencing, respectively. a, b Gene expressions of CgB-aat1, CgCbs, CgTyr, CgTyrp1 and CgTyrp2 in mantle by qPCR detection, respectively. The cDNA samples from EGFP-RNAi group (Egfp-i) and CgB-aat1 and CgCbs silencing group (B-aat1-i, Cbs-i) were analyzed by qPCR. The internal gene ef1α of C. giga was as control. All data were shown as mean ± SE (n = 6). Asterisk (*) was used to indicate the significant difference (P < 0.05). c Protein expressions of CgB-aat1, CgCbs, CgTyr, CgTyrp1 and CgTyrp2 in mantle by WB analysis, respectively. GAPDH was used as internal control
Effect of CgB-aat1 and CgCbs silencing on the expression of the key genes associated with melanin synthesis
The expression differences of CgCbs and key genes associated with melanin synthesis, CgTyr, CgTyrp1 and CgTyrp2 genes were analyzed by qPCR after the downregulation of CgB-aat1/CgCbs. As shown in Fig. 4a, b, CgCbs, participating in the intermediate process of pheomelanin synthesis, its expression was also decreased by 74.76% (P < 0.05) after CgB-aat1 silencing. And there was no significant change of CgB-aat1 gene expression in CgCbs interference group. CgTyr played a vital role in catalyzing melanin synthesis and its transcriptional level was significantly declined by 83.93% (P < 0.05) in CgCbs dsRNA feeding. However, no significant difference was observed after CgB-aat1 silencing. Tyrp1 and Tyrp2 were functioned in melanocyte migration and differentiation, CgTyrp1 and CgTyrp2 gene expression were also no obvious difference after CgTyrp1 and CgTyrp2 knockdown, respectively. The same phenomenon was occurred in corresponding protein expression (Fig. 4c).
Histology changes of mantle after RNA interference
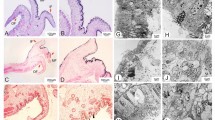
The effects of CgB-aat1 and CgCbs silencing on histological features of mantle were displayed in Fig. 5. Compared with NC group, brown-granule in the epithelial tissue of mantle was decreased in CgB-aat1 interference group (Fig. 5a). However, there was no obvious changes of melanin granules of mantle between CgCbs interference group and NC group.
Histology of mantle edge in CgB-aat1 and CgCbs silencing groups and control group. a1–a3 indicated the CgB-aat1 silencing group, b1–b3 meant the CgCbs silencing group, c1–c3 indicated the NC group (EGFP silencing group was used as negative control). Compared with NC group, the less brown granules (melanin granules) were observed in mantle edge after CgB-aat1 interference (a1–a3). And there was no significant change between CgCbs silencing group (b1–b3) and NC group (c1–c3). MG: melanin granules. Bars in a1–a3 and b1–b3 were 25 μm, in c1–c3 were 10 μm
Discussion
Skin color formation is an important physiological process in the life of organisms. The most abundant pigment is melanin, including eumelanin (brown/black) and pheomelanin (orange/red), have been the main research target in skin pigmentation so far. Previous researches verified that the biosynthesis of pheomelanin is inseparable from the participation of cysteine, which was transported to pigment cells by Slc7a11, followed conversed to pheomelanin by Cbs [1, 9]. It has been identified that Slc7a11 was an important regulator in pheomelanin production in the skin of mammals [6]. Nevertheless, little is known about the function of B-aat1 gene in invertebrate and we questioned whether it shares the similar function as Slc7a11 with regard to melanin synthesis. For this purpose, the deduced B-aat1 aa sequence possessed typical characteristics, which indicated that the processing of CgB-aat1 may be similar to Slc7a11 in other species. Besides, B-aat1 aa sequence also had several putative transmembrane domains, which was conserved with Slc7a11 aa sequences in red tilapia [9] and mammals [11,12,13]. Cbs is a downstream gene of Slc7a11 in pheomelanin biosynthesis and is also the key enzyme in transsulfuration pathway. The catalytic core of Cbs consist of a pyridoxal phosphate (PLP) domain containing the active site and CBS domains facilitating allosteric activation by S-adenosylmethionine [26]. Herein, like in other species, the CgCbs also include two domains (PALP and CBS), which stabilize the protein structure and enable the protein to play significant biological roles in the conversion of homocysteine to cysteine [27]. These findings indicated that structural and functional of B-aat1/Cbs in bivalves might be similar with Slc7a11/Cbs in other species.
Knowledge of temporal and tissue-specific expression characteristics of a gene is also worthy for predicting its function. Here, transcriptional level of CgB-aat1/CgCbs was both significantly up-regulated after D-veliger larvae and expression of CgCbs gene upped to the top at eyed-larva. Because eyed-larvae are the earliest development stage to develop the eye containing the pigment. This result indicated that CgB-aat1/CgCbs maybe function in preparation of melanin synthesis at early developmental stages. Moreover, the maximum expression level of CgB-aat1/CgCbs was both observed in central mantle, which acted on secretion of biomineralization proteins and pigmentation in the shell [28]. The gene expression level of CgB-aat1 was consistently high Slc7a11 in red tilapia and mammals [11, 12, 29,30,31], indicating that CgB-aat1 may also play an important role in color formation of shell color in mollusks. So far, there has been no report about the cellular location and expression of CgB-aat1 and CgCbs in other mollusks. IF results of CgB-aat1 and CgCbs protein was detected in cytoplasm of epithelial cell and nerve axons in mantle, where they may play vital roles in biochemical synthesis of pheomelanin. The same result was also happened in red tilapia [10]. Gasol et al. [32] discovered that Slc7a11/xCT is known as a multi-pass membrane protein, xCT maybe involved in membranous elements composition in the cytoplasm. Moreover, no more information on the cellular location of B-aat1 and Cbs is available in other mollusks.
Feeding of CgB-aat1-dsRNA and CgCbs-dsRNA resulted in significant down-regulation on CgB-aat1 and CgCbs mRNA as well as protein expression level, respectively. In vertebrate, Cbs also play a part in the pheomelanin biosynthesis under the activating of Slc7a11 expression [33]. Here, CgCbs gene expression also significantly suppressed after CgB-aat1 silencing. Histology result also showed that less brown-granules (melanin granules) were observed in CgB-aat1 silencing group. Affenzeller et al. [3] identified eumelanin and pheomelanin in Cepaea nemoralis. Taken together, the down-regulation of CgB-aat1 and CgCbs maybe reflected that the CgB-aat1- CgCbs axis in regulating the melanin synthesis pathway (specifically speaking pheomelanin) were existent in the Pacific oyster. However, mass spectrometric investigations for the presence of eumelanin and pheomelanin in different shell color of the Pacific oyster also need to be studied in the future.
In addition, the expressions result of Tyrp1 and Tyrp2 exhibited the lower expression level after CgCbs silencing, which is possible to effect melanin formation. CgTyr gene expression was also decreased under CgB-aat1 silencing and significantly down-regulated after CgCbs interference. Tyr can initiate catalyzed reaction and influence the rate of melanin synthesis [34]. As mentioned earlier, Tyr not only participate in eumelanin synthesis but also play an import role in pheomelanin synthesis pathway, in which it initiates biosynthesis of pheomelanin by providing dopaquinone for condensation with cysteine [1]. Combined the histological results, these studies reflected that CgB-aat1-CgTyr-CgCbs axis in pheomelanin regulation pathway maybe also conserved among C. gigas and other species, such as insect [35], fish [8, 36] and mammals [37]. Additionally, Emaresi et al. [38] suggested that the Tyr gene typically involved in eumelanin synthesis was strongly correlated and negatively associated with the Slc7a11 and Cbs genes typically involved in pheomelanin synthesis in the tawny owl. This research was slightly different with the results in C. gigas, indicating function in melanogenesis pathway study between B-aat1-Cbs and Tyr genes need to be research further.
Taken together, CgB-aat1-CgTyr-CgCbs axis maybe responsible for the pheomelanin synthesis and played an important role in mantle pigmentation of C. gigas. This finding provides a clue to the melanin pigment mechanisms in bivalves and will be beneficial to bivalve physiology in shell color formation. Nevertheless, the interplay between the eumelanin and pheomelanin synthesis pathway in C. gigas is still a problem that deserves further research.
Abbreviations
- B-aat1 :
-
B (0, +)-type amino acid transporter 1
- Cbs :
-
Cystathionine beta-synthase
- Ef1α :
-
α Subunit of elongation factor 1
- HPLC:
-
High-performance liquid chromatography
- HRP:
-
Horseradish peroxidase
- IF:
-
Immunofluorescence
- IPTG:
-
Isopropyl β-d-thiogalactoside
- Mitf :
-
Microphthalmia-associated transcription factor
- Pax3/7 :
-
Paired box 3/7
- Slc7a11 :
-
Solute carrier family 7 member 11 (xCT)
- SDS-PAGE:
-
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- Tyr :
-
Tyrosinase
- Tyrp1 :
-
Tyrosinase-related protein 1
- Tyrp2 :
-
Tyrosinase-related protein 2
- WB:
-
Western blot
References
Barek H, Sugumaran M, Ito S et al (2018) Insect cuticular melanins are distinctly different from those of mammalian epidermal melanins. Pigment Cell Melan Res 31(3):384–392. https://doi.org/10.1111/pcmr.12672
Williams ST (2017) Molluscan shell colour. Biol Rev 92(2):1039–1058. https://doi.org/10.1111/brv.12268
Affenzeller S, Wolkenstein K, Frauendorf H et al (2019) Eumelanin and pheomelanin pigmentation in mollusc shells may be less common than expected: insights from mass spectrometry. Front Zool 16:47–55. https://doi.org/10.1186/s12983-019-0346-5
True JR (2003) Insect melanism: the molecules matter. Trends Ecol Evol 18:640–647. https://doi.org/10.1016/j.tree.2003.09.006
Land EJ, Ramsden CA, Riley PA (2004) Quinone chemistry and melanogenesis. Methods Enzymol 378:88–109. https://doi.org/10.1016/S0076-6879(04)78005-2
Chintala S, Li W, Lamoreux ML et al (2005) Slc7a11 gene controls production of pheomelanin pigment and proliferation of cultured cells. Proc Natl Acad Sci USA 102(31):10964–10969. https://doi.org/10.1073/pnas.0502856102
Jiang Y, Zhang SH, Xu J et al (2014) Comparative transcriptome analysis reveals the genetic basis of skin color variation in common carp. PLoS ONE 9(9):e108200. https://doi.org/10.1371/journal.pone.0108200
Luo MK, Lu GQ, Yin HR et al (2021) Fish pigmentation and coloration: molecular mechanisms and aquaculture perspectives. Rev Aquac. https://doi.org/10.1111/raq.12583
Zhu W, Tian CX, Huang Y et al (2016) Comparative transcriptome analysis identifies candidate genes related to skin color differentiation in Red tilapia. Sci Rep 6:31347. https://doi.org/10.1038/srep31347
Wang LM, Bu HY, Song FB et al (2019) Characterization and functional analysis of slc7a11 gene, involved in skin color differentiation in the red tilapia. Comp Biochem Physiol A. https://doi.org/10.1016/j.cbpa.2019.110529
Tian X, Meng XL, Wang LY et al (2015) Molecular cloning, mRNA expression and tissue distribution analysis of Slc7all gene in alpaca (Lama paco) skins associated with different coat colors. Gene 555:88–94. https://doi.org/10.1016/j.gene.2014.10.029
He X, Li HT, Zhou ZY et al (2012) Production of brown/yellow patches in the SLC7A11 transgenic sheep via testicular injection of transgene. J Genet Genom 39(6):281–285. https://doi.org/10.1016/j.jgg.2012.04.005
Yang NS, Liu M, Zhao BH et al (2018) RNAi-mediated SLC7A11 knockdown inhibits melanogenesis-related genes expression in rabbit skin fibroblasts. J Genet 97(2):463–468. https://doi.org/10.1007/s12041-018-0945-5
Romero I, Tellez J, Yamanaka LE et al (2014) Transsulfuration is an active pathway for cysteine biosynthesis in Trypanosoma rangeli. Parasites Vectors 7:197. https://doi.org/10.1186/1756-3305-7-197
Lemer S, Saulnier D, Gueguen Y et al (2015) Identification of genes associated with shell color in the black-lipped pearl oyster Pinctada margaritifera. BMC Genom 16:568. https://doi.org/10.1186/s12864-015-1776-x
Yu FF, Qu BL, Lin DD et al (2018) Pax3 gene regulated melanin synthesis by tyrosinase pathway in Pteria penguin. Int J Mol Sci 19:3700. https://doi.org/10.3390/ijms19123700
Zhang SJ, Wang HX, Yu JJ et al (2018) Identification of a gene encoding microphthalmia-associated transcription factor and its association with shell color in the clam Meretrix petechialis. Comp Biochem Physiol B 225:75–83. https://doi.org/10.1016/j.cbpb.2018.04.007
Han ZQ, Li Q, Liu SK et al (2019) Genetic variability of an orange-shell line of the Pacific oyster Crassostrea gigas during artificial selection inferred from microsatellites and mitochondrial COI sequences. Aquaculture 508:159–166. https://doi.org/10.1016/j.aquaculture.2019.04.074
Li ZZ, Li Q, Liu SK et al (2021) Integrated analysis of coding genes and non-coding RNAs associated with shell color in the Pacific oyster (Crassostrea gigas). Mar Biotechnol 23:417–429. https://doi.org/10.1007/s10126-021-10034-7
Feng DD, Li Q, Yu H (2019) RNA interference by ingested dsRNA-expressing bacteria to study shell biosynthesis and pigmentation in Crassostrea gigas. Mar Biotechnol 21:526–536. https://doi.org/10.1007/s10126-019-09900-2
Li ZZ, Li Q, Xu CX et al (2022) Molecular characterization of Pax7 and its role in melanin synthesis in Crassostrea gigas. Comp Biochem Physiol B. https://doi.org/10.1016/j.cbpb.2022.110720
Du YS, Zhang LL, Xu F et al (2013) Validation of housekeeping genes as internal controls for studying gene expression during Pacific oyster (Crassostrea gigas) development by quantitative real-time PCR. Fish Shellfish Immunol 34:939–945. https://doi.org/10.1016/j.fsi.2012.12.007
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(T) (-Delta Delta C) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Liu HF, Wang J, Zhang LM et al (2021) Effects of recombinant AMH during oocyte maturation in spotted steed Hemibarbus maculatus. Aquaculture 543:736961. https://doi.org/10.1016/j.aquaculture.2021.736961
Li YJ, Ren LT, Fu HR et al (2021) Crosstalk between dopamine and insulin signaling in growth control of the oyster. Gen Comp Endocr 313:113895. https://doi.org/10.1016/j.ygcen.2021.113895
Koutmos M, Kabil O, Smith JL et al (2010) Structural basis for substrate activation and regulation by cystathionine beta-synthase (CBS) domains in cystathionine β-synthase. PNAS 107(49):20958–20963. https://doi.org/10.1073/pnas.1011448107
Giménez-Mascarell P, Majtan T, Oyenarte I et al (2018) Crystal structure of cystathionine β-synthase from honeybee Apis mellifera. J Struct Biol 202(1):82–93. https://doi.org/10.1016/j.jsb.2017.12.008
Addadi L, Joester D, Nudelman F et al (2006) Mollusk shell formation: a source of new concepts for understanding biomineralization processes. Chem Eur J 12(4):980–987. https://doi.org/10.1002/chem.200500980
Li HT, He X, Zhou ZY et al (2012) Expression levels of Slc7a11 in skin of kazakh sheep with different coat color. Hereditas 34:1314–1319. https://doi.org/10.3724/sp.j.1005.2012.01314
Li SJ, Wang C, Yu WH et al (2012) Identification of genes related to white and black plumage formation by RNA-Seq from white and black feather bulbs in ducks. PLoS ONE 7:e36592. https://doi.org/10.1371/journal.pone.0036592
Sato H, Tamba M, Kuriyama-Matsumura K et al (2000) Molecular cloning and expression of human xCT, the light chain of amino acid transport system xc−. Antioxid Redox Signal 2:665–671. https://doi.org/10.1089/ars.2000.2.4-665
Gasol E, Jimenez-vidal M, Chillaron J et al (2004) Membrane topology of system xc- light subunit reveals a re-entrant loop with substrate restricted accessibility. J Biol Chem 279(30):31228–31236. https://doi.org/10.1074/jbc.M402428200
Hoekstra HE (2006) Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity 97:222–234. https://doi.org/10.1038/sj.hdy.6800861
Busca R, Ballotti R (2000) Cyclic AMP a key messenger in the regulation of skin pigmentation. Pigment Cell Res 13:60–69. https://doi.org/10.1034/j.1600-0749.2000.130203.x
Sugumaran M (2009) Complexities of cuticular pigmentation in insects. Pigment Cell Melan Res 22(5):523–525. https://doi.org/10.1111/j.1755-148x.2009.00608.x
Fang WY, Huang JR, Li SZ et al (2022) Identification of pigment genes (melanin, carotenoid and pteridine) associated with skin color variant in red tilapia using transcriptome analysis. Aquaculture 547:737429. https://doi.org/10.1016/j.aquaculture.2021.737429
Morgan AM, Lo J, Fisher DE (2013) How does pheomelanin synthesis contribute to melanomagenesis? Two distinct mechanisms could explain the carcinogenicity of pheomelanin synthesis. Bioessays 35(8):672–676. https://doi.org/10.1002/bies.201300020
Emaresi G, Ducrest AL, Bize P et al (2013) Pleiotropy in the melanocortin system: expression levels of this system are associated with melanogenesis and pigmentation in the tawny owl (Strix aluco). Mol Ecol 22(19):4915–4930. https://doi.org/10.1111/mec.12438
Acknowledgements
This work was supported by the grants from National Natural Science Foundation of China (31972789), the China Agriculture Research System Project (CARS-49), and Earmarked Fund for Agriculture Seed Improvement Project of Shandong Province (2020LZGC016 and 2021LZGC027).
Author information
Authors and Affiliations
Contributions
QL conceived and designed the study. ZL, BH, LD and CH performed RNA interference experiment and collected the samples. ZL drafted the manuscript and analyzed the data, and QL revised the manuscript. ZL, BH, LD, CH and QL have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical approval
The Pacific oyster is neither an endangered nor protected species. All experiments in this study were conducted according to national and institutional guidelines.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Z., Hu, B., Du, L. et al. Involvement of B-aat1 and Cbs in regulating mantle pigmentation in the Pacific oyster (Crassostrea gigas). Mol Biol Rep 50, 377–387 (2023). https://doi.org/10.1007/s11033-022-08037-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-08037-1