Abstract
Background
Shikonin is a natural multipotent anti-tumorigenic compound. We investigated potential synergy between shikonin and anti-diabetic metformin against tumorigenic properties of breast cancer cell line MCF-7.
Methods and results
The IC50 of shikonin and metformin was determined after a single treatment of two cell lines MCF-7 and MDA-MB-231. We then measured optimal doses of each drug, used in combination, in MCF-7 cells. These sub-IC50 doses were co-applied for all subsequent combined treatments to evaluate their synergistic effects on MCF-7 tumorigenic properties. Next, we examined expression levels of the genes crucial for apoptosis, cell growth, and EMT using RT-PCR or real-time PCR and monitored CD44/CD24 ratios using flow cytometry. Binding energies between shikonin and growth molecules were measured by in silico simulation.
Shikonin caused significantly reduced cell survival that was accelerated by the synergizing presence of metformin. Drug combination induced apoptosis and ROS levels while fully blocking cell migration and reverting EMT. RT-PCR showed strong suppression of BCL-2 but induction of BAX and PTEN. Prolonged shikonin treatment caused a total loss of the nuclear membrane, whereas metformin prevented this damage while promoting apoptotic morphologies. Our real-time PCR detected reduced levels of EMT genes but increases in the anti-EMT gene CDH1. Combined treatment also reduced CD44/CD24 ratios in favor of chemosensitivity. Binding energies strongly favored shikonin interactions with growth-signaling molecules.
Conclusions
Shikonin and metformin synergize in inhibiting the tumorigenic activities of MCF-7 cells including their proliferation, invasiveness, and EMT with a potential to inhibit multidrug resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer (BC) occurs in the inner layer of mammary glands or lobules and ducts of the breast on a diverse causative ground of genetics/epigenetics, virus infection, immunity, family history, nutrition, and lifestyle [1]. Symptoms include a lump or mass in the affected breast tissue with changed breast shape or size, discharge from the nipple, breast rash and pain, and redbreast skin [1].
The World Health Organization considered BC the most common cancer as of 2021, as the disease accounted for 12% of all new annual cancer cases worldwide. Accordingly, BC continues to be the most commonly diagnosed cancer and the second leading cause of cancer deaths among U.S. women [2]. The rate of BC incidence among women shows no declining prospect and, in 2021, an estimated 281,550 new cases of invasive BC is expected to be diagnosed in women in the U.S., including 2650 new invasive cases, with a death toll that could rise to 43,600 affected women [3].
Despite tremendous advances in therapy management of BC and other cancers, multidrug resistance (MDR) remains the biggest challenge in fully eradicating the disease. In general, intra-tumoral heterogeneity and clonal evolution during tumor development form the foundations of cancer MDR [4] that develops inherently by tumor kinetics or is acquired in the course of tumor chemo- or radiotherapy [5]. Frequent chemotherapy promotes cancer MDR by several mechanisms. It upregulates drug transporter genes [6]. It also introduces post-treatment adaptive changes in cancer cell characteristics that consist of epithelial-mesenchymal transition (EMT) and its reversal to a cancer stem cell (CSC)-like phenotype or mesenchymal-epithelial transition (MET) [7]. Other effects of frequent chemotherapy include elevation of reactive oxygen species (ROS) that disrupts redox homeostasis and makes cancer cells more chemo-resistant via 'redox resetting' [8] and induction of CSCs to self-renew and differentiate into heterogeneous lineages of cancer cells [9] that actively contribute to tumor invasion, metastasis [10], and angiogenesis [11].
Effective therapeutic measures carefully designed to inhibit causative pathways of MDR are required in treating cancer. Targeted monotherapies may be ineffective for a vast majority of common tumors as these tumors are not dependent on a single “targetable” oncogenic activation and, over time, they can acquire resistance by activating multiple signaling pathway redundancies and adaptive mechanisms [12]. Updated technologies define targeted cancer therapy in the areas of small-molecule medicine and monoclonal antibodies. However, combination approaches that apply multiple therapeutic agents can either enhance toxicities or raise other side effects besides raising the financial costs of treatment. Targeted therapies based on genome editing approaches [13], on the other hand, are a novice to the clinic awaiting long-term trials for their net beneficial effects and may not offer better solutions than current monotargeting. This is why combination therapy using natural products is often in the spotlight. Cancer cell chemosensitivity is mechanistically enhanced only by co-inducing both apoptotic and non-apoptotic cell death, and this can be done by combined application of natural products [14].
To this end, we designed our current study to test the combined effect of anti-tumorigenic compound shikonin (SHKN) and anti-diabetic medicine metformin (MTFN). Shikonin is an active naphthoquinone isolated from the Chinese medicine Zi Cao [15] and exerts anti-cancer activities including cancer cell cycle arrest, induction of apoptosis and necroptosis, deregulation of cancer cell metabolism, and suppression of metastasis [15]. Metformin, on the other hand, essentially reduces the chance of cancer development by reducing blood glucose and inhibiting insulin and IGF-1 production [16]. Here, we compared the simultaneous versus the individual impact of SHKN and MTFN on BC cell morphology and expression of selected genes involved in cell survival, proliferation, migration, EMT, and CSC activities.
Material and methods
Cell culture and chemicals
Human BC cell lines MCF-7 and MDA-MB-231 were obtained from the National Cell Bank of Pasture Institute (Tehran, Iran) and cultured in Dulbecco’s modified eagle’s medium (DMEM; Gibco, 12,430,054) supplemented with 10% fetal calf serum (FBS; Gibco, A4766801), and maintained at 37 °C and 5% CO2. SHKN was purchased from Sigma (54,952–43-10) and dissolved in dimethyl sulfoxide (DMSO; Thermo Fisher, 20,688) and MTFN was gifted by Dr. Abidi Co (Tehran) and dissolved in PBS to form treatment stocks.
Cell death analyses
Inhibitory effects of varying dosages of SHKN and MTFN were measured using cell viability assay. MCF-7 and MDA cells were seeded in a 96-well plate at 12 × 103 cells per well overnight and treated with serial concentrations of SHKN, MTFN, or their combination. We applied SHKN in the range of 3 to 25 μM and MTFN from 3 to 70 mM. DMSO, PBS (drugs’ solvents), and untreated cells were used in parallel as our base controls. After a 24-h-incubation period, we exposed the cells to the MTT (Sigma, M5655) solution to measure viability. In the end, we read the absorbance at 580 nm using an ELISA reader. The calculated inhibitory concentrations (IC50s) represent the treatment concentrations that inhibit 50% of cells’ growth versus controls. For co-treatment and subsequent analyses, we used MCF-7 cells only.
Measurement of drug synergism
The experimental runs for SHKN-MTFN co-treatment of MCF-7 cells were designed using Design Expert Software (DOE) (Stat-Ease Inc., version 11). In accordance with the DOE, the optimum doses of the drugs must be established to eliminate the systematic error and reduce the number of experiments needed for analysis of the impact that several factors have on the multifaceted drug interactions. We employed the Central Composite Design (CCD), which is most commonly used with RSM, statistically a sound method of optimizing the parameters of a process needed for appropriate modeling. Therefore, we used RSM to predict the effects of independent variables (drugs) on each other and dependent variables (response; survival in this study) at five levels. Thirteen different runs were calculated to determine the optimal doses of drugs in combination, including 5 runs at the center point. To this end, a range of 1–8 μM SHKN and 10–30 mM MTFN were applied to keep lack of fit (LOF) insignificant. The obtained data were illustrated using a 3D-plot diagram to demonstrate the simultaneous effect of variables (drugs) on the response (cell viability).
Cell co-staining and live-cell count
Since acridine orange (AO) co-applied with ethidium bromide (EB) stains nuclear DNA green in live cells and orange in dead cells, we followed the standard AO/EB co-staining method to measure the percent of viable treated MCF-7 cells. First, a 3 mg/ml stock of ethidium bromide (Thermo Fisher, 15,585,011) in absolute ethanol and a 5 mg/ml stock of acridine orange (Thermo Fisher, A1301) in ethanol were prepared and stored in cold/dark. Next, equal volumes of the stock and PBS were mixed to constitute the working stain solution. Cell groups were suspended at a normal counting concentration from which 50 µL was mixed with 50 µL of the stain in 96-well plates and subjected to counting under fluorescent light. Images were captured using a fluorescence microscope coupled with a Nikon digital camera. Six random microscopic fields per well were selected to count an average of 120 cells counted for each cell group in triplicates (three wells per group). We counted green cells with the round clear nucleus as live cells, those green ones with bright green dots in their nuclei as early apoptotic, condensed and fragmented or disintegrated nuclei stained for orange as late apoptotic, and those orange-stained nuclei with uniform morphology as necrotic cells [17].
Reactive oxygen species measurement
We used Dichloro-dihydro-fluorescein Diacetate (DCFH-DA) (Thermo Fisher, D399) for ROS measurement. DCFH-DA is a fluorogenic dye that, upon cell uptake, is deacetylated by cellular esterases to a non-fluorescent compound, which is later oxidized by ROS into 2’-7’ dichlorofluorescein (DCF) [18]. MCF-7 cells were seeded 15 × 103 per well on 96-well plates 24 h before treatment with the IC50 of SHKN, MTFN, or their combination followed by 4–5 h incubation. The cells were re-fed with DMEM plus 5% FBS containing 10 μM DCFH-DA and incubated for 45 min at 37 °C. Detached cells were suspended in PBS to the final volume of 500 μL and analyzed for ROS levels using VARIAN Flow Cytometer (set at 495 nm and 500–550 nm excitation and emission wavelengths, respectively). In this experiment, we used untreated cell samples, DMSO-treated samples, also samples treated with DMSO + DCFH-DA as our negative controls, and H2O2-treated cells (2.5 µg/µL) stained with DCFH-DA as our positive control.
Evaluation of cell death by flow cytometry, Annexin V-FITC/PI double labeling
MCF-7 cells were seeded 4 × 105 per well in 6-well plates. After treatment with the individual or combined IC50 doses of the drugs for 24 h, they were harvested, PBS washed, and rinsed with cold PBS. Cell pellets were re-suspended with the stains and incubated for 15 min at 4 °C in dark before being analyzed by a FACSCalibur at 488 nm wavelength. A total of 100,000 cells were used per sample and the data were analyzed in FlowJo 7.6.1 software.
Flow cytometric analysis of stem cell surface markers CD24 and CD44
MCF-7 cells treated with IC50 doses of the drugs for 24 h were trypsinized and re-suspended in PBS. Fluorochrome-conjugated antibodies (1:1) against human CD24 (PE-conjugated) (Thermo Fisher, 12–0242-82) and CD44 (FITC-conjugated) (Thermo Fisher, 11–0441-82) were separately added to each cell suspension for mono-staining and incubated at 4 °C in dark for 30–40 min. The labeled cells were analyzed by flow cytometry to collect the final data.
Cell migration assay
Migration was assessed by a scratch assay. MCF-7 and MDA cells were seeded in 6-well plates (4 × 105 cells per well) in DMEM/F12 (GIBCO, A4192001) supplemented with 10% FBS, to become 80% confluent in 24 h. A sterile razor blade was used to scrape cells off the culture plate leaving a denuded area and a sharp visible demarcation line at the wound edge. After scraping the cell monolayer, the wounded monolayers were washed with PBS twice and were inspected immediately after wounding. Then sections of the wounds were selected according to the criteria, marked, and numbered. Next, the cells were incubated with single and combined concentrations of drugs for 6, 12, and 24 h (MCF-7) or 24 h only (MDA) and examined under phase-contrast microscopy. The photographs collected from the cell samples were trimmed and edited so to more closely represent the migration process. Migrated MCF-7 cells were counted in Sects. 500 μm in length, allowing a 20 μm space from the demarcation line to minimize the possible physical effects of cell movement resulting from cell proliferation. Statistical analysis was calculated by averaging a mean of six sections per test for each experiment. The number of migrated cells was expressed as mean ± SEM. The experiments were repeated at least three times in each group to assess reproducibility.
RNA extraction and reverse transcription (RT) PCR
RT-PCR was carried out as reported [19]. Briefly, total RNA was extracted, DNase treated and used (2 µg) for cDNA synthesis. The cDNA samples were first denatured at 95 °C for 2 min before amplifying target DNA fragments. Table 1 shows primer pairs plus cycling parameters applied for each gene candidate. Gel electrophoresis and band intensity measurements were duplicated as reported [19].
Real-time qPCR assay
The expression of some critical factors involved in migration and EMT/MET was quantified by quantitative real-time PCR. Each reaction included the cDNA template, target-specific primers, and SYBRGreen I PCR Master Mix (Thermo Fisher, 4,309,155). All results were normalized relative to the expression of human GAPDH and fold change expression was calculated by the delta-delta Ct method [20]. Human-specific primers and cycling conditions were shown in (Table 1).
Molecular docking
Molecular docking was used to estimate the inhibitory effects of SHKN on selected proteins involved in the main cell growth pathways. In the first step, the SHKN structure was taken from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/), and then, the optimized structure was obtained using the ATB webserver (Automated force field Topology Builder, http://compbio.biosci.uq.edu.au/atb). Subsequently, in the AutoDockTools (ADT) environment (http://autodock.scripps.edu/resources/adt), non-polar hydrogens were merged, and Gasteiger charges were added to the SHKN structure. Also, the number of rotatable bonds was considered. Next, 3D structures of the proteins were taken from the RCSB PDB database (https://www.rcsb.org/). Ligand molecules, nonessential chains, and water molecules, if present, were removed from each protein structure. Kollman charges and polar hydrogens were also added to protein structures. For each protein, a grid box was defined on the active site, and docking simulation was carried out by the use of AutoDuck Vina 1.1.2. Finally, Discovery Studio Visualization (DSV) was applied to visualize 2D and 3D binding structures.
Statistical analysis
The results are presented as mean ± SE. Statistical comparisons between groups were carried out using one-way ANOVA followed by the Student's t-test. We used data of triplicates or more repeats for each set of experiments, and comparisons were carried out only between relevant pairs of data. We considered a value of P < 0.05 as statistically significant, and P < 0.01 or P < 0.001 as highly significant.
Results
MCF-7 and MDA cell survival declines upon exposure to shikonin and metformin
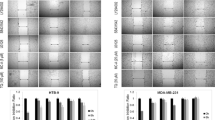
MCF-7 and MDA cells were treated with serial concentrations of SHKN and MTFN for 24 h before their viability was measured using MTT assay. The cells treated with higher concentrations of each drug appeared to be rounded up during exposure (Fig. 1A). As for MCF-7 cells, the MTT assay determined IC50 of SHKN 10 µM (Fig. 1B) and MTFN 60 mM (Fig. 1C). These figures for MDA cells stood at 17 µM for SHKN and 70 mM for MTFN. We also found that cell death is dose-dependent and accelerated beyond the range of the drug dose (Fig. 1B, C).
Reduced survival of MCF-7 and MDA-MB-231 cells treated with shikonin or metformin. A Morphological changes of the cells after treatment. DMSO was used as the solvent. Magnification: 40x. B and C represent the percent of survival after cell treatment, respectively, with increasing concentrations of shikonin and metformin. Statistical difference between each drug dose and its counterpart in control is shown by *, **, or ***
In silico analysis suggests SHKN-MTFN synergy in killing MCF-7 cells
To find the optimum doses of SHKN-MTFN combination for MCF-7 cells, we used Central Composite Design (CCD)-Response Surface Methodology (RSM) to make our analyses on 5 different surfaces (Suppl. Table 1). To select an effective range of drugs, an MTT assay was carried out and the results were used to design an experiment that included 13 tests (Suppl. Table 2). The RSM variances were statistically analyzed and, based on the results (Suppl Table 3), the most appropriate model was selected.
The RSM method designs a predicting model based on the compatibility of the data it produces and their interactions. According to RSM, the P < 0.05 and lack of fitness over 0.05 indicate that the selected model is statistically significant and highly capable of predicting response. A difference of < 0.2 between R2 and adjusted R2 is another indication of the model's correctness. In our study, we selected the second level model with P < 0.0001 and R2 = 0.9972 for our prediction purposes, where the difference between R2 and adjusted R2 was 0.002. Therefore, a graph was produced to represent different doses of drug combinations predicted by the model versus those examined in live cells (Fig. 2A) and predicted a quadratic model as the optimum model. According to this model, 7 µM (SHKN) and 13 mM (MTFN) as the optimum drug pair concentrations were selected. Drug-drug interactions were analyzed for SHKN and MTFN based on the 3D RSM graph (Fig. 2B) that indicated SHKN-MTFN interaction is statistically significant and the two drugs are functionally interrelated.
MCF-7 cell apoptosis is accelerated upon co-treatment with SHKN-MTFN
The AO/EB co-staining experiments revealed altered morphologies typical of apoptosis and/ or necrosis among cell samples (Fig. 3A). Compared to untreated controls, dying morphology appeared more distinctive in treatment intervals from 1 to 5 and 8 h. The most dominant morphology was atypical cell membrane at 5-h treatment and beyond that tended to be increasingly blebbing, and chromatin condensation and fragmented nucleus at 8 h. At 24 h treatment with SHKN alone, the membranes disappeared and an empty nucleus became visible so the cells appeared enucleated due to rapid disintegration by the drug before the stained could be absorbed (Fig. 3A; compare the inlets at the 24-h group with those at the other intervals). As appears from Fig. 3A, nuclear and cytoplasmic components coming out of the cells spread into the growth medium. The addition of MTFN to the cells prevented the loss of the nuclear membrane by SHKN and the cells displayed fragmented and punctated nuclei instead.
Induced apoptosis and necrosis among treated and co-treated MCF-7 cells. A Cell co-staining by Acridine orange and ethidium bromide followed by photography under the fluorescent microscope (Magnification: 20x). Closer presentation of changes is shown by inlets. B Percent of apoptotic cells. Each column in this and subsequent graphs represents an average of three independent experiments carries out in triplicates by random counting of microscopic fields as detailed in Methods. (± SEM; n = 3). C Flow cytometric analysis of treated and co-treated cell samples. D Percent of apoptotic and necrotic cells from (C). NEC, necrotic cells; E-AP, early apoptotic cells; L-AP, late apoptotic cells. In both (C) and (D) Symbols *, **, or *** indicate statistical differences between each column with its control counterpart. Symbols # and ‡ compare each column of combined treatment, respectively, with the shikonin-treated and metformin-treated columns, whereas symbol ζ shows differences between the two single treatments
Increased time of drug exposure from 1 to 8 and then to 24 h steadily increased the percent of apoptotic cells versus controls (Fig. 3B, P < 0.001 for SHKN and P < 0.05 for MTFN) but SHKN induced stronger rates of programmed cell death than did MTFN (P < 0.01). As for combined treatments with sub-IC50 doses of either drug, higher rates of apoptosis were induced compared to single treatments (P < 0.05 versus SHKN and P < 0.01 versus MTFN).
Further evidence indicating that both SHKN and MTFN induce apoptosis was provided after Annexin-V staining of treated MCF-7 cells followed by flow cytometry (Fig. 3C). Some necrotic cell death was also detected in SHKN-treated samples. Reduced rates of survival were more evident in co-treated samples versus single treatments. In fact, the co-treated cells showed the highest rates of both early and late apoptosis (Fig. 3D, P < 0.01 compared to controls and P < 0.05 compared to single treatments).
ROS levels are accelerated by SHKN-MTFN co-treatment of MCF-7 cells
We measured changes in ROS levels in our cell samples with 10 µM SHKN or 60 mM MTFN in single treatments and 7 µM SHKN and 13 mM MTFN in co-treatments. As shown in (Table 2), the intensity of DCFH-DA conversion to DCF as a sign of ROS elevation increased in SHKN-treated cells compared to controls, whereas MTFN alone had no significant effect on ROS levels. However, the increase by SHKN was further accelerated when measured in SHKN-MTFN co-treated cell samples compared to single treatments. Numerically, the intensity of adsorption at 520 nm wavelength increased from 78 units in MCF7 controls to 125 units in SHKN-treated and 211 units in co-treated samples (Table 2; P < 0.01).
Co-treated MCF-7 cells completely lose their migrating capacity
Cell treatment with either SHKN or MTFN significantly diminished cell migration and co-treated cells fully stopped migrating (Fig. 4A). As before, 10 µM SHKN or 60 mM MTFN were used for our single treatments and 7 µM SHKN and 13 mM MTFN in co-treatments. We quantified cell movement at serial time points of 6, 12, and 24 h post-treatment and concluded that while the reduction in movement by either drug reaches a maximum of 60%, cell movement was completely stopped in SHKN-MTFN co-treated cell samples (Fig. 4B; P < 0.01). Additionally, migration of our co-treated cells completely stopped at almost every time point we examined. Surprisingly, reduction of drug concentrations to half (3 µM SHKN and 7 mM MTFN) also fully stopped cell migration (data not shown). Besides MCF-7 cells, the anti-migration capacity of SHKN-MTFN synergy was tested and confirmed in MDA cells as well (Suppl. Figure 1).
Inhibition of MCF-7 cell migration by shikonin-metformin synergy. A Morphological changes in cell migration. We treated our cell samples with 10 µM shikonin and 60 mM metformin for individual treatments, and 7 µM shikonin plus 13 mM metformin for co-treatments. CTRL, DMSO-treated MCF-7 cells. The microscopic photographs were trimmed and edited so to more closely represent the migration process. Magnification: 10x. B Percent of migrating cells. The statistical difference between each column and controls is shown by*, ** or ***
Molecular mechanisms of drug action
Based on our morphological observations in single- and double-treated MCF-7 cells outlined above, we monitored cellular changes at molecular levels. Primarily, RT-PCR was performed on a few genes namely anti-apoptotic BCL-2, pro-apoptotic BAX, and tumor suppressor PTEN (Fig. 5A). SHKN-MTFN co-treatment significantly reduced BCL-2 levels but increased BAX and PTEN levels compared to both controls (Fig. 5A, P < 0.01) and single treatments (P < 0.05).
Altered expression of key genes by shikonin-metformin synergy. A RT-PCR products of cell samples treated with the same doses outlined in Fig. 4 were run on gel electrophoresis and images were collected. The intensity of the gel bands was measured as outlined in Method and expressed as a percent of mRNA relative to controls. B Relative mRNA levels of candidate molecules determined by real-time PCR experiments. Symbols * and ** show statistical differences between each column and its control counterpart, and # between cotreated samples and single treatments in the same group. C Flow cytometric analysis of changes in CD44/CD24 ratios by shikonin-metformin synergy. 1. untreated control cell samples, 2. shikonin-treated samples, 3. metformin-treated samples, 4. Co-treated samples. Each histogram represents either CD24 or CD44 mono-staining. The dotted histograms belong to the unstained samples, whereas the red-color histograms are stained for either antibody. The ratio for each group is shown on top of the relevant graph. D Simulation of binding between shikonin and AKT1 molecule. Supplement Fig. 2 shows images of shikonin binding to additional key molecules. See text for explanation and Table 3 for binding energies. (Color figure online)
Following the full blockage of cell migration by SHKN-MTFN co-treatment, we speculated that the co-treatment could block and even reverse the EMT. To test this hypothesis, we examined the expression levels of pro-and anti-EMT genes in our test samples using real-time PCR analyses. SHKN-treated and MTFN-treated MCF-7 cells showed significantly reduced levels of pro-EMT genes SNAIL, MMP9, CTNNB1, WNT1, and STAT3 whereas levels of anti-EMT CDH1 significantly increased in both groups (Fig. 5B; P < 0.05). More importantly, the SHKN-MTFN co-treated cell group showed further diminished expression of pro-EMT genes compared to controls (P < 0.01) and single treatments (P < 0.05). Also, significantly accelerated levels of CDH1 expression were detected in co-treatment samples compared to both controls (P < 0.01) and single treatments (P < 0.05).
Next, we analyzed CSC markers by flow cytometry. SHKN (10 µM) and MTFN (40 mM) reduced CD44/CD24 ratio from 154 folds in controls, respectively, to 22 folds and 36.5 folds (Fig. 5C; P < 0.05). This ratio was reduced to 1.7 folds when the two drugs were co-applied, indicating that both compounds inhibit cancer stem cell marker (CD44) while maximum inhibition is achieved by their combination.
These observations encouraged us to gain insight into the mechanism of SHKN interactions with some intracellular molecules. As previously reported [21], the KEGG pathway analysis nominates signaling pathways PI3K/AKT and MAPK/ERK that are commonly active in BC and, therefore, considered potential SHKN targets. We captured the following protein structures from Protein Data Bank (PDB) for our docking analyses: PI3K, AKT1/2, ERK1/2, mTOR, MEK1/2, RAF1, HRAS. Following the preparation of ligands and proteins, docking analyses were carried out in silico to simulate drug-receptor bindings (Fig. 5D and Suppl Fig. 2). Figure 5D represents the simulation of extensive binding between shikonin and AKT1 as one of the most commonly mutated genes in various cancer types. Suppl Fig. 2 shows images of shikonin binding to additional key molecules. As shown in (Table 3), the binding energy between SHKN and each target molecules we analyzed was below − 6 kcal/Mol indicating a strong binding. We collected the information for five conformers of each molecule and found similar binding energy in each case (data not shown).
Discussion
Combination therapy induces drug-drug interactions that create synergistic anti-cancer effects and elevate the drug sensitivity of cancer cells by targeting the sources of resistance such as CSCs and mitochondrial ROS accumulation [22, 23]. A clear example comes from ER− BC cells in which SHKN breaks taxol resistance and re-sensitizes the cells to the drug by preventing tyrosine kinases from activity [24]. Our combined approach revealed the net effects of SHKN and MTFN together on cancer cell biology and morphology. It also showed that the sub-lethal doses of the two compounds in combination are sufficient to exert a sound anti-cancer effect. Indeed, SHKN-MTFN joint activities on cancer cell fate were compatible with what we predicted by virtual analysis that indicated the two drugs synergize without compromising one another’s effectiveness.
Effective genotoxicity of drugs is achievable by targeting cancer cells in culture dishes or, at most, animal models of tumors. Minimized doses may compromise SHKN efficacy but, in parallel, reduce the chance of damaging normal cells. In a separate study to be completed, we tested the notion that a combination of SHKN and MTFN in reduced doses potentiates chemosensitivity of tumor cells while maintaining the efficacy of SHKN toxicity specifically against cancer cells. Similarly, our current study found that reduced doses of the two in combination do not compromise the anti-tumorigenic properties observed with higher doses of either drug when applied individually.
The alliance between SHKN and MTFN significantly reduced MCF-7 viability and accelerated cell death. Parallel with increases in the timeframe of drug exposure, morphological changes typical of apoptosis were detected in cell shape, including membrane blebbing and nuclear disintegration. We observed a complete loss of nucleus in the AO/EB co-stained cells after 24 h treatment with SHKN, but not with shorter exposures and that MTFN prevented these changes from occurring. MTFN-mediated nuclear preservation might be due to progerin downregulation. Reports about disease conditions indicate that the accumulation of progerin, a toxic form of Lamin A, in the nuclear membrane and within the nucleus distorts nuclear architecture and negatively affects nuclear processes, leading to accelerated cellular aging and premature senescence [25]. The progerin-encoding gene LMNA is induced by SRSF1 [26]. MTFN reduces progerin levels by inhibiting SRSF1, thereby protecting the nuclear membrane from progerin-mediated damages [27].
Our flow cytometry data indicated SHKN significantly induces apoptosis and, to some extent, necrosis. However, induction of either cell death mechanism by MTFN alone was not as significant, whereas the combination of the two drugs strikingly accelerated both phenomena. SHKN induces cell death via both p53-dependent and independent mechanisms [28, 29], including apoptosis, necrosis, and autophagy. Yeh, et al. (2015) showed that SHKN, at low concentrations, induces apoptosis and senescence, whereas its higher concentrations induce both apoptosis and necrosis [28]. Compatible with our flow cytometry findings, RT-PCR showed combined SHKN-MTFN treatment causes more significant BCL-2 downregulation and more PTEN and Bax upregulation than either drug alone does.
SHKN caused more strong cell migration inhibition than MTFN. In comparison, SHKN-MTFN synergy completely stopped the cells from migrating. Furthermore, our sequential time point experiments showed that the cells crossing the scratch line are not the products of new proliferation activities since MCF-7 doubling time is 30–40 h. Also, detection of full migration inhibition at early time points implies that the phenomenon did not occur due to reduced cell viability post-treatment either.
We detected similar patterns of migration inhibition in MDA-MB-231, a triple-negative aggressive BC (TNBC) cell line. Recently Bao et al. (2021) reported SHKN-mediated inhibition of migration in MDA-MB-231 and BT549 (both TNBC) occurs via PTEN interactions with miR-17-5p [30]. Our co-treated MCF-7 cells carried induced levels of PTEN expression. We also showed SHKN strongly binds AKT1. Similar to the report by Bao et al., these changes are in favor of migration inhibition.
Shikonin effect on cell migration is mediated by its impact on genetic pathways that regulate cytoskeleton formation, including microtubule dysfunction [31]. Cell migration is halted by inhibition of cytoskeleton development and reversal of EMT. STAT3 binds to and inhibits stathmin, a protein that accelerates depolymerization of microtubules by binding to tubulin subunits [32]. Both SHKN and MTFN contribute to migration inhibition by downregulating STAT3 [33, 34]. Therefore, complete migration inhibition detected in our co-treated cells likely occurred by SHKN-MTFN synergy.
The anti-migration effects of MTFN are also mediated by diminished ROS levels and reduced COX2 expression [35]. SHKN induces apoptosis [36] and necroptosis [37] via upregulation of ROS, whereas MTFN reduces endogenous ROS production by its preventative activities [38]. SHKN-mediated ROS elevation in our MCF-7 cells was significant, whereas MTFN did not cause tangible changes. Surprisingly, the combination of the two compounds significantly accelerated ROS production. This indicates SHKN-MTFN synergy on migration inhibition is a net effect of both the same and different mechanisms of action that the two drugs exert on the cell. For example, Marinello et al. (2019) reported that MTFN can increase BC cell sensitivity to doxorubicin-induced oxidative stress [39].
The observations outlined above matched with upregulation of anti-EMT CDH1and downregulation of pro-EMT genes SNAIL, STAT3, MMP9, and CTNNB1, detected by our real-time PCR. These molecules actively contribute to the cellular movement by regulating cell adhesion and cell-surface attachment or detachment. Our study limited the examination of the EMT to these molecules within their long list. Given that EMT proteins are strongly regulated by posttranslational modifications, a new dimension to our RNA-level study could be to examine changes at post-translation levels and by adding new EMT-related molecules to the list.
We used CD44/CD24 ratio as a reliable marker for characterizing CSCs. The consistency of the CD44/CD24 ratio from primary tumors to circulating tumor cells (CTCs) and distant metastases has been validated [40]. The study also found that high CD44/CD24 is an indication of tumor malignancy. Interactions between CSC marker CD44 and its ligand hyaluronan increase resistance to numerous drugs [41] and stimulates expression of ABCG2 and ABCB1, the main drug transporters involved in BC drug resistance [42, 43]. Given the role of CD44 in cell growth and drug resistance [44], we examined the notion that SHKN-MTFN synergy can minimize the CD44/CD24 ratio, as an indicator of MDR. Han et al. reported the anti-drug resistance properties of SHKN [45]. In contrast, Wu et al. showed SHKN partially induces drug resistance in cancer cells [46]. On the other hand, MTFN is a candidate compound for adjuvant therapy of drug-resistant tumors [47]. Both SHKN and MTFN significantly reduced the CD44/CD24 ratio in our MCF-7 cells, indicating declined levels of CD44 expression. More importantly, SHKN-MTFN synergy accelerated this decline and, therefore, could be employed as a potent inhibitor of drug resistance in cancer cells.
Our last but least attempt in gaining insight into the SHKN mechanism of anti-growth action examined its binding to growth signaling molecules that often undergo mutations and deregulations in BC. The selected molecules consisted of several key elements of PI3K/AKT and MAPK/ERK, two main growth signaling pathways. Similar to our study on astaxanthin [21], the current in-silico examinations found strongly favorable binding energies exist between SHKN and several key signaling molecules. These virtual observations support our current in vitro findings, although they require future expansion and experimental validation.
In conclusion, our study produced a broad image of apoptotic events induced by SHKN-MTFN synergy in MCF-7 cells: SHKN-MTFN combination, on one hand, accelerates cancer cell apoptosis, in essence by combining their pro-PTEN/Bax and anti-BLC2 activities. On the other hand, it elevates ROS levels, thereby inhibiting cell migration and reverting EMT. These events, together with a sharp decline in CD44/CD24 ratios, indicate the high potential of the SHKN-MTFN combination to abrogate chemoresistance and irreversibly induce mechanisms of apoptotic and necrotic cell deaths.
The future prospect that lies ahead of our study could potentially expand our initial establishment of the SHKN-MTFN synergy. Next steps could potentially test SHKN-MTFN co-effect on several cell lines of defined profile from BC and other cancer types, examine many more EMT genes, and evaluate CSC markers by co-staining them in single samples compared to separate examinations. Molecular changes we tested at RNA levels can be re-visited more specifically to monitor post-translational modifications. Experimental validation of our virtual findings about SHKN binding to intracellular signaling molecules add a new dimension to these studies that together could be tested on patient tumor samples and in vivo models of resistance to candidate chemodrugs.
Data availability
Any materials and data are available from the corresponding author on reasonable request.
Code availability
Not applicable.
References
Breast Cancer Treatment. National Cancer Institute. Updated 8 Apr, 2021. https://www.cancer.gov/types/breast/patient/breast-treatment-pdq#_125. Accessed 5 Jan 2022
DeSantis CE, Fedewa SA, Goding Sauer A, Kramer JL, Smith RA, Jemal A (2016) Breast cancer statistics, 2015: convergence of incidence rates between black and white women. CA Cancer J Clin 66:31–42
U.S. Breast Cancer Statistics. https://www.breastcancer.org/symptoms/understand_bc/statistics. Accessed 5 Jan 2022
Burrell RA, Swanton C (2014) Tumour heterogeneity and the evolution of polyclonal drug resistance. Mol Oncol 8:1095–1111. https://doi.org/10.1016/j.molonc.2014.06.005
Vasan N, Baselga J, Hyman DM (2019) A view on drug resistance in cancer. Nature 575:299–309. https://doi.org/10.1038/s41586-019-1730-1
Ichihashi N, Kitajima Y (2001) Chemotherapy induces or increases expression of multidrug resistance-associated protein in malignant melanoma cells. Br J Dermatol 144:745–750. https://doi.org/10.1046/j.1365-2133.2001.04129.x
Shibue T, Weinberg RA (2017) EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol 14:611–629. https://doi.org/10.1038/nrclinonc.2017.44
Liu Y, Li Q, Zhou L et al (2016) Cancer drug resistance redox resetting renders a way. Oncotarget 7:42740–42761
Phi LTH, Sari IN, Yang Y-G et al (2018) Cancer Stem Cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment. Stem Cells Int 2018:5416923. https://doi.org/10.1155/2018/5416923
Li S, LI Q, (2014) Cancer stem cells and tumor metastasis. Int J Oncol 44:1806–1812. https://doi.org/10.3892/ijo.2014.2362
Zhao Y, Bao Q, Renner A et al (2011) Cancer stem cells and angiogenesis. Int J Dev Biol 55:477–482. https://doi.org/10.1387/ijdb.103225yz
Basmadjian C, Zhao Q, Bentouhami E, Djehal A, Nebigil CG, Johnson RA, Serova M, de Gramont A, Faivre S, Raymond E, Désaubry LG (2014) Cancer wars: natural products strike back. Front Chem 2:20. https://doi.org/10.3389/fchem.2014.00020
Zhan T, Rindtorff N, Betge J et al (2019) CRISPR/Cas9 for cancer research and therapy. Semin Cancer Biol 55:106–119. https://doi.org/10.1016/j.semcancer.2018.04.001
Yuan R, Hou Y, Sun W et al (2017) Natural products to prevent drug resistance in cancer chemotherapy: a review. Ann N Y Acad Sci 1401:19–27. https://doi.org/10.1111/nyas.13387
Guo C, He J, Song X et al (2019) Pharmacological properties and derivatives of shikonin—A review in recent years. Pharmacol Res 149:104463. https://doi.org/10.1016/j.phrs.2019.104463
Dowling RJ, Goodwin PJ, Stambolic V (2011) Understanding the benefit of metformin use in cancer treatment. BMC Med 9:33. https://doi.org/10.1186/1741-7015-9-33
Kasibhatla S, Amarante-Mendes GP, Finucane D, Brunner T, Bossy-Wetzel E, Green DR (2006) Acridine Orange/ethidium bromide (AO/EB) staining to detect apoptosis. CSH Protoc. https://doi.org/10.1101/pdb.prot4493
Eruslanov E, Kusmartsev S (2010) Identification of ROS using oxidized DCFDA and flow-cytometry. Methods Mol Biol 594:57–72. https://doi.org/10.1007/978-1-60761-411-1_4
Gardaneh M, Gholami M, Maghsoudi N (2011) Synergy between glutathione peroxidase-1 and astrocytic growth factors suppresses free radical generation and protects dopaminergic neurons against 6-hydroxydopamine. Rejuvenation Res 14:195–204. https://doi.org/10.1089/rej.2010.1080
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Gardaneh M, Nayeri Z, Akbari P et al (2020) Molecular simulations identify target receptor kinases bound by astaxanthin to induce breast cancer cell apoptosis. Arch Breast Cancer. https://doi.org/10.32768/abc.20207272-82
Wang T, Narayanaswamy R, Ren H, Torchilin VP (2016) Combination therapy targeting both cancer stem-like cells and bulk tumor cells for improved efficacy of breast cancer treatment. Cancer Biol Ther 17:698–707. https://doi.org/10.1080/15384047.2016.1190488
Fiorillo M, Tóth F, Sotgia F, Lisanti MP (2019) Doxycycline, Azithromycin and Vitamin C (DAV): A potent combination therapy for targeting mitochondria and eradicating cancer stem cells (CSCs). Aging (Albany NY) 11:2202–2216
Li W, Liu J, Jackson K et al (2014) Sensitizing the therapeutic efficacy of taxol with shikonin in human breast cancer cells. PLoS ONE 9:e94079. https://doi.org/10.1371/journal.pone.0094079
Goldman RD, Shumaker DK, Erdos MR et al (2004) Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci 101:8963–8968. https://doi.org/10.1073/pnas.0402943101
Lopez-Mejia IC, Vautrot V, De Toledo M et al (2011) A conserved splicing mechanism of the LMNA gene controls premature aging. Hum Mol Genet 20:4540–4555. https://doi.org/10.1093/hmg/ddr385
Egesipe AL, Blondel S, Lo Cicero A, Jaskowiak AL, Navarro C, Sandre-Giovannoli A, Levy N, Peschanski M, Nissan X (2016) Metformin decreases progerin expression and alleviates pathological defects of Hutchinson-Gilford progeria syndrome cells. NPJ Aging Mech Dis 2:16026. https://doi.org/10.1038/npjamd.2016.26
Yeh YC, Liu TJ, Lai HC (2015) Shikonin induces apoptosis, necrosis, and premature senescence of human A549 lung cancer cells through upregulation of p53 expression. Evid Based Complement Alternat Med 2015:620383. https://doi.org/10.1155/2015/620383
Wang F, Mayca Pozo F, Tian D et al (2020) Shikonin inhibits cancer through P21 upregulation and apoptosis induction. Front Pharmacol 11:861. https://doi.org/10.3389/fphar.2020.00861
Bao C, Liu T, Qian L et al (2021) Shikonin inhibits migration and invasion of triple-negative breast cancer cells by suppressing epithelial-mesenchymal transition via miR-17-5p/PTEN/Akt pathway. J Cancer 12:76–88. https://doi.org/10.7150/jca.47553
Wiench B, Eichhorn T, Paulsen M, Efferth T (2012) Shikonin directly targets mitochondria and causes mitochondrial dysfunction in cancer cells. Evidence-Based Complement Altern Med 2012:726025. https://doi.org/10.1155/2012/726025
Verma NK, Dourlat J, Davies AM et al (2009) STAT3-Stathmin interactions control microtubule dynamics in migrating t-cells. J Biol Chem 284:12349–12362. https://doi.org/10.1074/jbc.M807761200
Trinh SX, Nguyen HTB, Saimuang K et al (2017) Metformin inhibits migration and invasion of cholangiocarcinoma cells. Asian Pac J Cancer Prev 18:473–477
Cao HH, Liu DY, Lai YC, Chen YY, Yu LZ, Shao M, Liu JS (2020) Inhibition of the STAT3 signaling pathway contributes to the anti-melanoma activities of shikonin. Front Pharmacol 11:748. https://doi.org/10.3389/fphar.2020.00748
Schexnayder C, Broussard K, Onuaguluchi D, Poché A, Ismail M, McAtee L, Llopis S, Keizerweerd A, McFerrin H, Williams C (2018) Metformin inhibits migration and invasion by suppressing ROS production and COX2 expression in MDA-MB-231 breast cancer cells. Int J Mol Sci 19(11):3692. https://doi.org/10.3390/ijms19113692
Chang I-C, Huang Y-J, Chiang T-I et al (2010) Shikonin induces apoptosis through reactive oxygen species/extracellular signal-regulated kinase pathway in osteosarcoma cells. Biol Pharm Bull 33:816–824. https://doi.org/10.1248/bpb.33.816
Zhang Z, Zhang Z, Li Q et al (2017) Shikonin induces necroptosis by reactive oxygen species activation in nasopharyngeal carcinoma cell line CNE-2Z. J Bioenerg Biomembr 49:265–272. https://doi.org/10.1007/s10863-017-9714-z
Algire C, Moiseeva O, Deschênes-Simard X et al (2012) Metformin reduces endogenous reactive oxygen species and associated DNA damage. Cancer Prev Res 5:536–543. https://doi.org/10.1158/1940-6207.CAPR-11-0536
Marinello PC, Panis C, Silva TNX et al (2019) Metformin prevention of doxorubicin resistance in MCF-7 and MDA-MB-231 involves oxidative stress generation and modulation of cell adaptation genes. Sci Rep 9:5864. https://doi.org/10.1038/s41598-019-42357-w
Li W, Ma H, Zhang J, Zhu L, Wang C, Yang Y (2017) Unraveling the roles of CD44/CD24 and ALDH1 as cancer stem cell markers in tumorigenesis and metastasis. Sci Rep 7(1):13856. https://doi.org/10.1038/s41598-017-14364-2.Erratum.In:SciRep.2018Mar6;8(1):4276
Toole BP (2009) Hyaluronan-CD44 interactions in cancer: paradoxes and possibilities. Clin Cancer Res 15:7462–7468. https://doi.org/10.1158/1078-0432.CCR-09-0479
Misra S, Ghatak S, Toole BP (2005) Regulation of MDR1 expression and drug resistance by a positive feedback loop involving hyaluronan, phosphoinositide 3-kinase, and ErbB2. J Biol Chem 280:20310–20315. https://doi.org/10.1074/jbc.M500737200
Gilg AG, Tye SL, Tolliver LB et al (2008) Targeting Hyaluronan Interactions in Malignant Gliomas and Their Drug-Resistant Multipotent Progenitors. Clin Cancer Res 14:1804–1813. https://doi.org/10.1158/1078-0432.CCR-07-1228
Chen C, Zhao S, Karnad A, Freeman JW (2018) The biology and role of CD44 in cancer progression: therapeutic implications. J Hematol Oncol 11:64. https://doi.org/10.1186/s13045-018-0605-5
Han W, Li L, Qiu S et al (2007) Shikonin circumvents cancer drug resistance by induction of a necroptotic death. Mol Cancer Ther 6:1641–1649. https://doi.org/10.1158/1535-7163.MCT-06-0511
Wu H, Xie J, Pan Q et al (2013) Anticancer agent shikonin is an incompetent inducer of cancer drug resistance. PLoS ONE 8:e52706. https://doi.org/10.1371/journal.pone.0052706
Coyle C, Cafferty FH, Vale C, Langley RE (2016) Metformin as an adjuvant treatment for cancer: a systematic review and meta-analysis. Ann Oncol 27:2184–2195. https://doi.org/10.1093/annonc/mdw410
Funding
This study was financially supported by a grant from National Institute of Genetic Engineering and Biotechnology (NIGEB) (Grant 589).
Author information
Authors and Affiliations
Contributions
ART carried out most of the experiments. PG designed and implemented a part of experiments including invasion and cell co-staining plus molecular docking. FS contributed to the dose determination of shikonin and metformin as well as editing and reviewing the manuscript. MG was a grantee of the project, supervised the first author, and designed the whole structure of the manuscript in detail.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tabari, A.R., Gavidel, P., Sabouni, F. et al. Synergy between sublethal doses of shikonin and metformin fully inhibits breast cancer cell migration and reverses epithelial-mesenchymal transition. Mol Biol Rep 49, 4307–4319 (2022). https://doi.org/10.1007/s11033-022-07265-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07265-9