Abstract
Bacteria, fungi, virus and nematode constitute the primary class of pathogens causing plant diseases. Plant–pathogen interactions are crucial for the identification of the host and pathogen and further establishments of a network of interaction that can cross regulate the gene expressions in both sides. After infection, the correct identification of pathogen through various molecular interactions elicit a defense response against the pathogen by alteration of gene expression by the host. Co-evolution of pathogen gives them the ability to counter the virulence response of the host and pathogen can also modulate the host gene expression pattern to make it more susceptible to the infection. Small non-coding RNA molecules (siRNAs and miRNAs) efficiently modulate gene expression at the transcriptional and post-transcriptional level and play a vital role in host defense. The pathogen can also use this double-sided sward in their defense by deregulating the plant immunity via transcriptional control of plant genes utilizing RNA interference or suppressing the host RNA interference response with the help of various RNA silencing suppressor proteins. This mini-review focused on the miRNAs involvement in host defense and how different families of these non-coding regulatory RNAs regulate the defense response against the pathogen.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants survival against the pathogen depends upon a wide variety of factors that governs the plant immunity. Over the period of time, plant also evolves to develop intricate mechanisms that recognize the various surface molecular patterns present in the pathogens. These pathogen associated molecular patterns (PAMPs) when recognized by plant cell elicits a strong defense response known as PAMP triggered immunity (PTI). However due to continuous random mutations, many plant pathogens have evolved a diverse variety of effector proteins that can inhibit PTI signaling when come in contact with the plant cell [1]. In order to impede with such pathogens, plants have developed another type of immunity known as effector triggered immunity (ETI). This second layer of inducible defense uses resistance proteins to counteract with the effectors. The resistance proteins coding genes are being triggered by small RNAs (sRNAs) by their differential regulation contributing another class of defense mechanism to plants [2]. Small RNAs in plants are able to regulate development, stress tolerance and antiviral defenses [3]. These small RNAs namely microRNAs (miRNAs) and small interfering RNAs (siRNAs) provide immunity to plants against several infections by inducing RNA interference (RNAi) pathways that trigger transcriptional gene silencing.
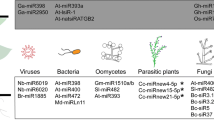
As the small RNAs present in plants contribute to plant immunity, in the same way the pathogen-derived sRNAs regulate the virulence of pathogens. It is remarkable about these small RNAs that their regulation is not limited to the specific organism in which they are synthesized. If inserted into any other interacting species, they trigger silencing of genes in the host organism, also known as Cross Kingdom RNAi. This phenomenon is exhibited by a specific range of pathogens and parasites to suppress the host plant gene through host induced gene silencing (HIGS) method. The engineered plants are developed that triggers RNAi against various pathogens to generate an immune response [4]. These small RNA populations that participate in developing immune response against the pathogen consist of mainly micro RNAs and small interfering RNAs.
MicroRNAs (miRNA) are short non-coding regulatory RNAs of 20–22 nucleotides long that participate in various cellular processes such as in development, proliferation and stress response at the post-transcriptional level. They bind to the 3′ untranslated region (UTR) of the target mRNA to control the expression of their target mRNAs. Many miRNAs can bind with a single UTR due to availability of multiple binding sites or multiple sites for a single miRNA that suggests a complex post-transcriptional control of gene expression exerted by these regulatory RNAs.
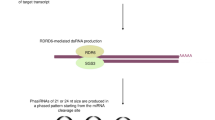
The processing of miRNAs is done from the primary transcript using a two-step sequential mechanism involving two RNase III nucleases. The miRNAs produced either from the processing of a host intron or by transcription from their dedicated promoters. Firstly, the nuclear RNase III processes primary precursor (pri-miRNA) into an approximately 70 nucleotide long stem-loop structure. The microprocessor complex contains Drosha. The cleavage of the 5′ and 3′ ends of the pri-miRNA by the two RNase domains of Drosha determines the length of pre-miRNA. A complex of Exportin-5 and Ran-GTP exports resultant pre-miRNA to the cytoplasm. Dicer helps with the final maturation of miRNA and another RNase III nuclease processes the pre-miRNA into a 22 bp double-stranded RNA. The formation of ribo-nucleoprotein complex known as RISC (miRNA-Induced Silencing Complex) is coupled with the processing of miRNA. The RISC minimally contains one strand of the miRNA which is called as “guide strand”. In addition to Dicer, TRBP, PACT and Argonaute (Ago) proteins are also involved. The silencing is executed by the engaging of the complex with the target while the passenger strand, another strand of the miRNA is generally digested [5].
Small interfering RNAs are similar to miRNAs in size but differ in precursor structure, biogenesis pathway and modes of action. In plants, four types of siRNAs are found, namely ta-siRNA, fa-siRNA, nat-siRNA and hc-siRNA. These small RNAs are known to provide disease resistance to plants [6]. However in the present mini-review, the focus will be on miRNAs. During plant–pathogen interaction, a plethora of these miRNAs get involved and shows differential expression.
Micro RNAs in fungal–plant interaction
A wide variety of phytopathogenic fungi is known to attack plants to fulfill their nutritional requirements. Several miRNAs have been identified which are known to provide critical resistance during the fungal attack by interacting with several regions of R genes. The plant host miRNAs participate in disease modulation upon fungal infection either by up or down regulation. Plant miRNAs modulate phytohormones homeostasis by regulating the expression of their target transcripts. The miR408 acts as a negative regulator of plantacyanins and laccase that plays a key role in cell to cell signaling, lignin formation and stress response in Cicer arietinum seedlings and plants [7]. Similarly, the susceptible and resistant wheat cultivars infected by Puccinia graminis at early and late phase of infection show differential regulation of miR408 [8]. Likewise, the upregulation of Osa-miR7695 provides enhanced resistance against blast fungus in rice [9] thus proving a potential biomarker for miRNA based detection of the Magnaporthe oryzae disease in resistant and non-resistant Vietnamese rice cultivars [10].
Most of the miRNAs that are activated by the fungus target several genes at once and each of these genes controls a cascade of other pathways that regulate the whole cellular process. Thus a detailed understanding of disease pathogenesis can be achieved by understanding the regulation and crosstalk of gene expression during pathogen infection and pathological development. In many cases, same genes are being targeted by different miRNAs but their expression is dependent upon the plant and pathogen being studied. For instance, the miRNA family (miR482) was reported to regulate the expression of Nucleotide Binding Site-Leucine Rich Repeats (NBS-LRRs), a major class of specific pathogen resistance genes (R-genes). When young potato plants were infected with Verticillium dahlia, the miR482e expression was found to be downregulated and several Nucleotide Binding Site-Leucine Rich Receptor targets of miR482e were upregulated [11]. The decreased expression of miR482e is a counter defense strategy of plant against the pathogen and in transgenic plants when miR482 expression is upregulated, the susceptibility of plants towards the fungus infection increases. In case of Phytophthora infestans infection, an upregulation of miR482/2118 in early stage of infection cause increased susceptibility of tomato to the pathogen [12].
On infection with Verticillium dahliae, over 65 miRNA families found to exhibit an altered expression in two cotton cultivars [13]. The scientist also identified three cotton specific miRNAs-Ptc-miR482, Ptc-miR1444, and Ptc-miR1448 which were able to cleave the Poly Phenol Oxidase (PPO) and disease resistance protein genes. The downregulation of these specific miRNAs during the initial Verticillium infection caused an upregulation of PPO and disease resistance gene, thus providing an enhanced resistance towards the fungus [14]. Another mode of Verticillium-cotton disease signaling was also found in a report, where miR166 and miR159 expression was upregulated targeting Ca2+-dependent cysteine protease (Clp-1) and an isotrichodermin C-15 hydroxylase (HiC-15) that are essential for fungal virulence [15]. The expression of miRNAs also differs in different fungus parts. MicroRNAs identified from hyphae and micro-conidia of Fusarium oxysporum (Fon) shows differential expression in these two propagules [16]. The study also revealed down-regulation of Fon-miR7696a-3p and Fon-miR6108a leading to an increased biosynthesis of toxin related gene expression.
MicroRNAs are also involved in targeting various transcriptional factors under pathogen attack. Out of 12 miRNAs that get upregulated in Populus trichocarpa under infection from canker pathogen, miR156 found to target TC-rich repeats and Wi-box motif [17]. In case of Fusarium oxysporum infection in Persicaria minor, a class of miRNAs- miR156b, miR172a, miR319, miR858 and miR894 were found to target transcriptional factors that are involved in biotic stress response and plant development [18]. MicroRNAs also enhance transcriptional priming in defense response. During Magnaporthe oryzae infection in rice, miR7695 down-regulate the expression of natural resistance-associated macrophage protein 6 (OsNramp6) [19]. NRAMP6 is associated with the transport of iron in rice. As such the plants cultivated under high iron content showed resistance against the fungus. MicroRNAs also inhibit the MAP kinase-mediated immunity in plants under fungal attack. Wang et al. [20] reported that under Fusarium oxysporum attack in cotton, miR5272 limits the plant immunity through downregulation of MAPKK6 expression thus increasing the susceptibility towards disease.
In a recent in-silico study that aims in identification and comparative analysis of different miRNAs and their targets in wheat under powdery mildew, leaf rust and blast disease revealed that the functionality of 18 different miRNAs depends entirely on the specificity of hosts [21]. The researchers also observed target multiplicity and selective gene targeting caused due to multiple miRNAs.
Phytohormones also play a major role in plant defense against the pathogen attack. During plant–fungal interaction, the pathogen mimics the function of phytohormones which enables them to manipulate the regulation of signaling in plant defense resulting in hormonal imbalance and inapt defense response [22]. Plants also adapt to downregulate the auxin synthesis when attacked to obstruct the pathogen from using host hormones as a virulence factor. MicroRNA also regulates the key components of hormonal signaling pathway. During powdery mildew infection in wheat, auxin-mediated response got enhanced on downregulating the transport inhibitor response 1 (TIR1) with an upregulation of miR393 [23]. Therefore, regulation of hormonal pathways by miRNA can be considered as essential targets for transgene engineering during plant–fungal interaction.
Micro RNAs in bacteria–plant interaction
Bacteria always have been a prominent agent that leads to biotic stress in plants by causing various diseases. Besides many other mechanisms through which plant-counter attack the pathogen attack, RNA silencing seems to be a custom made defense mechanism against bacteria. During bacterial–plant interaction, many miRNAs get involved that manipulates auxin, abscisic acid and jasmonic acid signaling pathways and thus regulates the plant defense mechanism. Navarro et al. [24] was first to report that miR393 regulate the plant immunity against Pseudomonas syringae in Arabidopsis. The miR393 induced by bacterial elicitor flg22, targets TIR1, AFB2 and AFB3 mRNAs and suppressed the auxin signaling pathway. Apart from miR393, another class of micro RNAs-miR160, miR167 and miR390 also regulate the auxin signaling and inhibiting Pseudomonas syringae growth in Arabidopsis [25]. Thus, during the course of evolution, plants have developed intricate counter-attack mechanism including mi-RNA mediated gene regulation that can suppress multiple components of the auxin signaling pathways in response to bacterial infection. Auxin being a prominent phytohormone enhances apical growth which provides nutrition (carbon and nitrogen) to the developing pathogen thus increasing their susceptibility and virulence. Moreover, auxin also suppress salicylic acid-mediated defense pathway in plants and in turn aids in increasing the pathogenesis [26].
Apart from auxin, a wide class of miRNAs also targets the signaling pathways of other hormones such as Abscisic acid (ABA), Jasmonic acid (JA) and Salicylic acid (SA) which are involved in anti-bacterial defense mechanism in plants. During Pseudomonas infection in Arabidopsis, a total of 20 differentially regulated miRNA families were observed that directly or indirectly targets the genes involved in signaling pathways of ABA, JA and SA [26]. These actions suggested that instead of targeting plants immune system the miRNAs helps in fine tuning of defense responses. MicroRNAs are also shown to regulate Pathogen-associated molecular patterns triggered immunity (PAM-PTI). In transgenic Arabidopsis, the overexpression of miR160a enhanced PAMP-induced callose deposition, thus adding a layer in plant defense while miR398b and miR773 reduce the deposition showing a complex interconnection between two classes of miRNAs [27]. Moreover, the type of bacteria interacting with the plants also affects the expression levels of miRNAs. Tobacco plants infected with Agrobacterium tumefaciens shows a higher expression of nta-miR393, whereas a higher expression of nta-miR167 was observed when plants were infected with Bacillus subtilis [28]. However, the increased expression of nta-miR393 and nta-miR167 also increases the expression of various flavonoid derivatives.
Small RNAs in viral–plant interaction
Immunity against viruses is mainly provided by Post Transcriptional Gene Silencing (PTGS) as viruses are obligate intracellular pathogens that depend on the plant for maintaining their life cycle. At the time of infection, viruses introduce their DNA or RNA into the host either in double stranded or single stranded form and PTGS get triggered to arrest virus replication and spreading in the plant. Recognition of PTGS was done in case of transgenics in Potato virus X (pvx) infection. The double stranded RNA of virus triggers the formation of various virus derived small interfering RNAs (VsiRNAs) by DCLs such as Cucumber yellows clostero virus (CuYV), Cucumber mosaic virus (CMV), Turnip mosaic potyvirus (TuMV), Tomato yellow leaf curl virus (TYLCV) and Watermelon mosaic virus (WMV) [29]. These VsiRNAs are attached with AGOs complex to direct DNA or RNA silencing. The activity of RDRs promotes production of secondary VsiRNAs and supports the systemic silencing [30]. The accomplishment of anti viral immunity is achieved by RNA silencing which can be seen as the utilization of dependency of viral life cycle on the host. In counter defense, viruses have also developed the strategy inhibiting host’s RNA silencing mechanism and counteracting the plant immunity with a variety of proteins known as viral suppressors of RNA silencing (VSRs) [31]. An efficient VSR can distinguish between an infected and a healthy plant host. Some VSRs are also known to target plant genes responsible for immune response.
During plant–virus interactions a diverse array of virus-responsive small RNAs has been found to be involved. On infection with Turnip mosaic virus (TuMV), two classes of miRNAs bra-miR158 and bra-miR1885 highly specific to Brassica rapa were greatly upregulated [32]. These class of miRNAs targeted Toll/Interleukin receptor domain-containing (TIR)–Nucleotide‐binding site (NBS)—Leucine‐rich repeat (LRR)—disease resistance (R) protein.
Viroids are the smallest (250–400 nt) known pathogens that are naked, single stranded circular RNAs molecules and replicates inside the nucleus or chloroplast. Viroids are known to infect plants and also initiate a series of small RNAs. Full methylation of the Potato spindle tuber viroid (PSTVd) cDNA sequence in PSTVd-infected tissues is carried out by the viroid-induced RNA silencing and RdDM [33]. Later, it was established that the RNA silencing is targeted and activated by viroids in infected potato plants. Citrus exocortis viroid (CEVd) after replication generates positively polarized 50-phosphorylated and 30-methylated vdsiRNAs. The placing of most of the CEVd Viroid-related siRNAs (vdsiRNAs) is within the right-end domain [34]. Some viroids like Chrysanthemum chlorotic mottle viroid (CChMVd) and Avocado sun blotch viroid (ASBVd) replicate in the chloroplast. The production of vdsiRNAs from negative and positive polarities is triggered by PLMVd and CChMVd [35]. The vdsiRNAs produced by ASBVd in leaves exhibit infection symptoms [36]. Hence, Avsunviroidae and Pospiviroidae, both viroid families, are capable of generating vdsiRNAs in different plants [37]. The generation of vdsiRNAs from both the positive and the negative strands of the viroid genome highlight the cardinal processing of vdsiRNAs from the viroid genomic RNAs secondary structure. A general mechanism of miRNAs role in different pathogenic attack against plants is illustrated in Fig. 1.
Micro RNAs in nematode–plant interaction
Plant parasitic nematodes are microscopic thread-like roundworms that survive in a wide range of environment that causes severe damage to crops. Sedentary endoparasitic nematodes are known to be the most damaging plant–parasitic nematodes (PPNs) [38]. These are categorized into two main groups—the root-knot nematodes (RKNs) of the genus Meloidogyne and the cyst nematodes (CNs) of the genera Heterodera and Globodera [39]. These parasites have a tendency to prick into roots of the host and induce the formation of specialized feeding structures which provide the resources needed for the development of nematode. Giant multinucleate feeding cells are formed due to the differentiation of five to seven root cells which is induced by Root-knot nematodes whereas cyst nematode triggers the formation of a multinucleate syncytium. An extensive reprogramming of gene expression in the targeted root cells is involved in the formation of the feeding cells by nematodes [40].
The attack of nematodes in plants is reciprocated by the expression of myriads of miRNAs in root cells of various plant species. The correct feeding site formation requires certain modules containing miRNAs and transcription factors that are targeted by nematodes. The studies on Arabidopsis thaliana during nematodal attack have shown that the formation of the feeding sites manipulate a number of plant functions including cytoskeleton, cell wall modification, cell cycle, plant defense and phytohormone pathways [41].
The RKN induces miRNAs which are involved in the gall formation in Arabidopsis that dissects galls and uninfected roots [42]. This approach helped in identifying 62 miRNAs responsible for inducing galls upon infection by Meloidogyne javanica at 3 dpi (dots per inch), and 24 miRNAs by Meloidogyne incognita at 7 and/or 14 dpi. Only two miRNAs were found to express same profile in three stages of gall formation: upregulation of miR390, and repression of miR319. On analyzing one or more of the five developmental stages of the tomato galls, 17 miRNAs were identified [43] whereas in whole cotton roots, 16 miRNAs were identified at 10 dpi upon infection by M. incognita [44]. The comparative studies of susceptible and resistant tomato cultivars revealed five RKN-responsive miRNAs in the jasmonic acid-deficient spr2 mutant at 3 dpi [45]. There are some conserved miRNA families which are present in similar expression profiles in galls from different plant species at similar time points. For instance, in Arabidopsis, tomato, and cotton galls the conserved miR159 is upregulated at 10 to 14 dpi, and miR172 is upregulated at 3 to 4 dpi in A. thaliana and tomato [46].
Identification of 30 mature differentially expressed (DE) miRNAs in Arabidopsis syncytia through sequencing showed that these miRNAs are induced by Heterodera schachtii at 4 and 7 dpi. A latest study of syncytia from tomato plants infected with Globodera rostochiensis performed at 3, 7, and 10 dpi, led to the identification of 200 to 300 miRNAs at each stage as DE [47]. The CN Heterodera glycine also promotes the expression of many miRNAs families upon infection. On analyzing RNA libraries from susceptible and resistant varieties of soyabean upon infection with soyabean CN, a total of 60 miRNAs from 25 miRNAs families were found to be DE [48]. Some miRNAs are upregulated in resistant lines relative to susceptible lines whereas some are downregulated. A comparative study of the expression profiles of conserved miRNAs in response to CN infection led to identification of some miRNAs as DE, with the same expression profile, in several plant species. The roots of Arabidopsis infected by H. schachtii showed down regulation of miR396b and the miR167 family at 4 and 7 dpi [32] and in Solanum lycopersicum, G. rostochiensis induces syctia at 3 and 7 dpi [49].
MicroRNAs actively contribute in the providing immunity against nematodal attack either by the variations in expressing of plant miRNAs which is directly affected by the nematode or caused due to alteration of plant hormonal cycle. Most studies until now have emphasized on miRNAs, but few studies principally based on the siRNAs have shown that the roots of Arabidopsis infected with PPNs have elucidated an overexpression of 24 nt siRNAa associated with RNA-directed DNA methylation in galls [50]. At last, cross-kingdom RNAi can happen while nematodes are interacting with plants. Conclusions can only be drawn if small RNAs from both side of the interaction are studied unified. The nematode-induced miRNAs are over-expressed and silence their corresponding targets. This might help in extracting important information about plant–nematode interaction and development of crop plants with nematode resistance. Table 1 enlist some recent studies on the action of miRNAs in plant defense mechanism.
Conclusions
In addition to playing an important role in development, growth and nutrition, sRNAs constitute a central part of plant immunity against the plant pathogens. As evident from the research that RNAi can work in a cross-kingdom manner, this double-sided sword can be used by host and pathogen both to regulate the gene expression of each other in a tough of war manner. The miRNAs can modulate the expression of various disease-associated resistance gene after the infection and can also bind to the transcriptional factors to produce a robust immune response. Phytohormones such as auxins, ascorbic acid are critically important for the healthy growth and nutrition are also up or down-regulated by modulating their associated signaling pathways utilizing miRNA and is an efficient strategy to generate resistance against pathogen. Virus derived miRNA can specifically inhibit viral function by transcriptional inhibition of viral mRNA. Some well-adapted viruses can also evade this RNAi mediated resistance of plants by suppressing it using many RNAi suppressors. Nematode infection requires a specified modification at the site of infection by reprogramming the gene expression, and the miRNA can modulate this reprogramming to elicit a defense response against the nematode. As miRNA can targets multiple mRNAs by binding them in a partial complementary manner, thus making them useful for creating resistance against many plant diseases and transgenic plants can be designed with improved resistance to various pathogens.
References
Nurnberger T, Kemmerling B (2018) Pathogen-associated molecular patterns (PAMP) and PAMP‐triggered immunity. Annu Plant Rev 6:16–47. https://doi.org/10.1002/9781119312994.apr0362
Wu L, Chen H, Curtis C, Fu ZQ (2014) Go in for the kill: how plants deploy effector-triggered immunity to combat pathogens. Virulence 5(7):710–721. https://doi.org/10.4161/viru.29755
Kamthan A, Chaudhuri A, Kamthan M, Datta A (2015) Small RNAs in plants: recent development and application for crop improvement. Front Plant Sci 6:208. https://doi.org/10.3389/fpls.2015.00208
Zotti M, dos Santos EA, Cagliari D, Christiaens O, Taning CNT, Smagghe G (2018) RNA interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest Manage Sci 74(6):1239–1250. https://doi.org/10.1002/ps.4813
Weiberg A, Jin H (2015) Small RNAs—the secret agents in the plant–pathogen interactions. Curr Opin Plant Biol 26:87–94. https://doi.org/10.1016/j.pbi.2015.05.033
Shahid S, Kim G, Johnson NR, Wafula E, Wang F, Coruh C, Bernal-Galeano V, Phifer T, Depamphilis CW, Westwood JH, Axtell MJ (2018) MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 553(7686):82–85. https://doi.org/10.1038/nature25027
Romo S, Labrador E, Dopico B (2001) Water stress regulated gene expression in Cicer arietinum seedlings and plants. Plant Physiol Biochem 39(11):1017–1026. https://doi.org/10.1016/S0981-9428(01)01318-3
Gupta OP, Sharma P, Gupta RK, Sharma I (2014) Current status on role of miRNAs during plant–fungus interaction. Physiol Mol Plant Pathol 85:1–7. https://doi.org/10.1016/j.pmpp.2013.10.002
Campo S, Peris-Peris C, Siré C, Moreno A, Donaire L, Zytnicki M, Notredame C, Llave C, SanSegundo B (2013) Identification of a novel microRNA (miRNA) from rice that target an alternatively spliced transcript of the Nramp6 (Natural resistance‐associated macrophage protein6) gene involved in pathogen resistance. New Phytol 199(1):212–227. https://doi.org/10.1111/nph.12292
Quoc NB, Phuong NDN, Trang HTT, Phi NB, Chau NNB (2019) Expression of osa-miR7695 against the blast fungus Magnaporthe oryzae in Vietnamese rice cultivars. Eur J Plant Pathol 155(1):307–317. https://doi.org/10.1007/s10658-019-01772-5
Yang L, Mu X, Liu C, Cai J, Shi K, Zhu W, Yang Q (2015) Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J Integr Plant Biol 57(12):1078–1088. https://doi.org/10.1111/jipb.12348
de Vries S, Kukuk A, von Dahlen JK, Schnake A, Kloesges T, Rose LE (2018) Expression profiling across wild and cultivated tomatoes supports the relevance of early miR482/2118 suppression for Phytophthora resistance. Proc R Soc Biol Sci 285(1873):20172560. https://doi.org/10.1098/rspb.2017.2560
Yin Z, Li Y, Han X, Shen F (2012) Genome-wide profiling of miRNAs and other small non-coding RNAs in the Verticillium dahliae–inoculated cotton roots. PLoS ONE 7(4):e35765. https://doi.org/10.1371/journal.pone.0035765
Jagannadham PTK, Muthusamy SK, Chidambaranathan P (2019) Micromics: a novel approach to understand the molecular mechanisms in plant stress tolerance. In: Wani SH (ed) Recent approaches in omics for plant resilience to climate change. Springer, New York, pp 93–108
Zhang T, Zhao YL, Zhao JH, Wang S, Jin Y, Chen ZQ, Fang YY, Hua CL, Ding SW, Guo HS (2016) Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nature Plants 2(10):1–6. https://doi.org/10.1038/nplants.2016.153
Jiang X, Qiao F, Long Y, Cong H, Sun H (2017) MicroRNA-like RNAs in plant pathogenic fungus Fusarium oxysporum f. sp. niveum are involved in toxin gene expression fine tuning. 3Biotech 7(5):354. https://doi.org/10.1007/s13205-017-0951-y
Zhao JP, Jiang XL, Zhang BY, Su XH (2012) Involvement of microRNA-mediated gene expression regulation in the pathological development of stem canker disease in Populus trichocarpa. PLoS ONE 7(9):e44968. https://doi.org/10.1371/journal.pone.0044968
Samad AFA, Ali NM, Ismail I, Murad AMA (2016) Analysis of miRNAs targeting transcription factors in Persicaria minor induced by Fusarium oxysporum. AIP Conf Proc 1784(1):020009. https://doi.org/10.1063/1.4966719
Sanchez-Sanuy F, Peris-Peris C, Tomiyama S, Okada K, Hsing YI, San Segundo B, Campo S (2019) Osa-miR7695 enhances transcriptional priming in defense responses against the rice blast fungus. BMC Plant Biol 19(1):1–16. https://doi.org/10.1186/s12870-019-2156-5
Wang C, He X, Wang X, Zhang S, Guo X (2017) ghr-miR5272a-mediated regulation of GhMKK6 gene transcription contributes to the immune response in cotton. J Exp Bot 68(21–22):5895–5906. https://doi.org/10.1093/jxb/erx373
Nair MM, Krishna TS, Alagu M (2020) Bioinformatics insights into microRNA mediated gene regulation in Triticum aestivum during multiple fungal diseases. Plant Gene 21:100219. https://doi.org/10.1016/j.plgene.2019.100219
Fonseca S, Radhakrishnan D, Prasad K, Chini A (2018) Fungal production and manipulation of plant hormones. Curr Med Chem 25(2):253–267. https://doi.org/10.2174/0929867324666170314150827
Nowara D, Gay A, Lacomme C, Shaw J, Ridout C, Douchkov D, Hensel G, Kumlehn J, Schweizer P (2010) HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22(9):3130–3141. https://doi.org/10.1105/tpc.110.077040
Navarro L, Jay F, Nomura K, He SY, Voinnet O (2008) Suppression of the microRNA pathway by bacterial effector proteins. Science 321(5891):964–967. https://doi.org/10.1126/science.1159505
Zhang W, Gao S, Zhou X, Chellappan P, Chen Z, Zhou X, Zhang X, Fromuth N, Coutino G, Coffey M (2011) Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol Biol 75:93–105
Mutka AM, Fawley S, Tsao T, Kunkel BN (2013) Auxin promotes susceptibility to Pseudomonas syringae via a mechanism independent of suppression of salicylic acid-mediated defenses. Plant J 74(5):746–754. https://doi.org/10.1111/tpj.12157
Li Y, Zhang Q, Zhang J, Wu L, Qi Y, Zhou JM (2010) Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol 152(4):2222–2231. https://doi.org/10.1104/pp.109.151803
Nazari F, Safaie N, Soltani BM, Shams-Bakhsh M, Sharifi M (2017) Bacillus subtilis affects miRNAs and flavanoids production in Agrobacterium–Tobacco interaction. Plant Physiol Biochem 118:98–106. https://doi.org/10.1016/j.plaphy.2017.06.010
Miao B, Yang GS, Chen W, Lin RM, Jian L, Mao ZC, Xie BY (2016) Characterization and function of Tomato yellow leaf curl virus-derived smallRNAs generated intolerant and susceptible tomato varieties. J Integr Agric 15:1785–1797. https://doi.org/10.1016/S2095-3119(15)61315-6
Burgyán J, Havelda Z (2011) Viral suppressors of RNA silencing. Trends Plant Sci 16(5):265–272. https://doi.org/10.1016/j.tplants.2011.02.010
Stavolone L, Prigigallo MI, Fabrizio C (2020) Plant viruses against RNA silencing-based defenses: strategies and solutions. In: Poltronieri P, Hong Y (eds) Applied plant biotechnology for improving resistance to biotic stress. Academic Press, Boca Raton, pp 225–250
Hewezi T, Howe P, Maier TR, Baum TJ (2008) Arabidopsis small RNAs and their targets during cyst nematode parasitism. Mol Plant Microbe Interact 21(12):1622–1634. https://doi.org/10.1094/MPMI-21-12-1622
Dalakouras A, Dadami E, Bassler A, Zwiebel M, Krczal G, Wassenegger M (2015) Replicating Potato spindle tuber viroid mediates de novo methylation of an intronic viroid sequence but no cleavage of the corresponding pre-mRNA. RNA Biol 12(3):268–275. https://doi.org/10.1080/15476286.2015.1017216
Byrne D, Grzela R, Lartigue A, Audic S, Chenivesse S, Encinas S, Claverie JM, Abergel C (2009) The polyadenylation site of Mimivirus transcripts obeys a stringent ‘hairpin rule.’ Genome Res 19(7):1233–1242. https://doi.org/10.1101/gr.091561.109
St-Pierre P, Hassen IF, Thompson D, Perreault JP (2009) Characterization of the siRNAs associated with peach latent mosaic viroid infection. Virology 383(2):178–182. https://doi.org/10.1016/j.virol.2008.11.008
López-Carrasco MA, Flores Pedauye R (2017) The predominant circular form of avocado sunblotch viroid accumulates in plant as a free RNA adopting a rod-shaped secondary structure unprotected by tightly bound host proteins. J Gen Virol 98(7):1913–1922. https://doi.org/10.1099/jgv.0.000846
Hammann C, Steger G (2012) Viroid-specific small RNA in plant disease. RNA Biol 9(6):809–819. https://doi.org/10.4161/rna.19810
Blok VC, Jones JT, Phillips MS, Trudgill DL (2008) Parasitism genes and host range disparities in biotrophic nematodes: the conundrum of polyphagy versus specialisation. BioEssays 30:249–259. https://doi.org/10.1002/bies.20717
Jones JT, Haegeman A, Danchin EGJ, Gaur HS, Helder J, Jones MGK (2013) Top 10 plant–parasitic nematodes in molecular plant pathology. Mol Plant Pathol 14:946–961. https://doi.org/10.1111/mpp.12057
Li X, Wang X, Zhang S, Liu D, Duan Y, Dong W (2012) Identification of soybean microRNAs involved in soybean cyst nematode infection by deep sequencing. PLoS ONE 7(6):e39650. https://doi.org/10.1371/journal.pone.0039650
Gheysen G, Mitchum MG (2019) Phytoparasitic nematode control of plant hormone pathways. Plant Physiol 179:1212–1226. https://doi.org/10.1104/pp.18.01067
Cabrera J, Olmo R, Ruiz-Ferrer V, Abreu I, Hermans C, Martinez-Argudo I (2018) A phenotyping method of giant cells from root-knot nematode feeding sites by confocal microscopy highlights a role for chitinase-like 1 in Arabidopsis. Int J Mol Sci 19:429. https://doi.org/10.3390/ijms19020429
Kaur P, Shukla N, Joshi G, Vijaya Kumar C, Jagannath A, Agarwal M (2017) Genome-wide identification and characterization of miRNAome from tomato (Solanum lycopersicum) roots and root-knot nematode (Meloidogyne incognita) during susceptible interaction. PLoS ONE 12:1–25. https://doi.org/10.1371/journal.pone.0175178
Pan X, Nichols RL, Li C, Zhang B (2019) MicroRNA-target gene responses to root knot nematode (Meloidogyne incognita) infection in cotton (Gossypium hirsutum L.). Genomics 111:383–390. https://doi.org/10.1016/j.ygeno.2018.02.013
Zhao W, Li Z, Fan J, Hu C, Yang R, Qi X, Chen H, Zhao F, Wang S (2015) Identification of Jasmonic acid-associated microRNAs and characterization of the regulatory roles of the miR319/TCP4 module under root-knot nematode stress in tomato. J Exp Bot 66(15):4653–4667. https://doi.org/10.1093/jxb/erv238
Lei P, Han B, Wang Y, Zhu X, Xuan Y, Liu X, Fan H, Chen L, Duan Y (2019) Identification of MicroRNAs that respond to soybean cyst nematode infection in early stages in resistant and susceptible soybean cultivars. Int J Mol Sci 20(22):5634. https://doi.org/10.3390/ijms20225634
Koter MD, Święcicka M, Matuszkiewicz M, Pacak A, Derebecka N, Filipecki M (2018) The miRNAome dynamics during developmental and metabolic reprogramming of tomato root infected with potato cyst nematode. Plant Sci 268:18–29. https://doi.org/10.1016/j.plantsci.2017.12.003
Tian B, Wang S, Todd TC, Johnson CD, Tang G, Trick HN (2017) Genome-wide identification of soybean microRNA responsive to soybean cyst nematodes infection by deep sequencing. BMC Genomics 18:1–13. https://doi.org/10.1186/s12864-017-3963-4
Święcicka M, Skowron W, Cieszyński P, Dąbrowska-Bronk J, Matuszkiewicz M, Filipecki M (2017) The suppression of tomato defence response genes upon potato cyst nematode infection indicates a key regulatory role of miRNAs. Plant Physiol Biochem 113:51–55. https://doi.org/10.1016/j.plaphy.2017.01.026
Ruiz-Ferrer V, Cabrera J, Martinez-Argudo I, Artaza H, Fenoll C, Escobar C (2018) Silenced retrotransposons are major rasiRNAs targets in Arabidopsis galls induced by Meloidogyne javanica. Mol Plant Pathol 19:2431–2445. https://doi.org/10.1111/mpp.12720
Sun L, Lin C, Du J, Song Y, Jiang M, Liu H, Zhou S, Wen F, Zhu C (2016) Dimeric artificial microRNAs mediate high resistance to RSV and RBSDV in transgenic rice plants. Plant Cell Tissue Organ Cult 126(1):127–139. https://doi.org/10.1007/s11240-016-0983-8
Soto-Suárez M, Baldrich P, Weigel D, Rubio-Somoza I, San Segundo B (2017) The Arabidopsis miR396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens. Sci Rep 7:44898. https://doi.org/10.1038/srep44898
Liang C, Hao J, Li J, Baker B, Luo L (2019) Artificial microRNA-mediated resistance to cucumber green mottle mosaic virus in Nicotiana benthamiana. Planta 250(5):1591–1601. https://doi.org/10.1007/s00425-019-03252-w
Santos LS, Maximiano MR, Megias E, Pappas M, Ribeiro SG, Mehta A (2019) Quantitative expression of microRNAs in Brassica oleracea infected with Xanthomonas campestris pv. campestris. Mol Biol Rep 46(3):3523–3529. https://doi.org/10.1007/s11033-019-04779-7
Yu X, Gong H, Cao L, Hou Y, Qu S (2020) MicroRNA397b negatively regulates resistance of Malus hupehensis to Botryosphaeria dothidea by modulating MhLAC7 involved in lignin biosynthesis. Plant Sci 292:110390. https://doi.org/10.1016/j.plantsci.2019.110390
Cui J, Jiang N, Hou X, Wu S, Zhang Q, Meng J, Luan Y (2020) Genome-wide identification of lncRNAs and analysis of ceRNA networks during tomato resistance to Phytophthora infestans. Phytopathology 110(2):456–464. https://doi.org/10.1094/PHYTO-04-19-0137-R
Cui C, Wang JJ, Zhao JH, Fang YY, He XF, Guo HS, Duan CG (2020) A Brassica miRNA regulates plant growth and immunity through distinct modes of action. Mol Plant 13(2):231–245. https://doi.org/10.1016/j.molp.2019.11.010
Ramachandran SR, Mueth NA, Zheng P, Hulbert SH (2020) Analysis of miRNAs in two wheat cultivars infected with Puccinia striiformis f.sp. tritici. Front Plant Sci 10:1574. https://doi.org/10.3389/fpls.2019.01574
Zhang Q, Li Y, Zhang Y, Wu C, Wang S, Hao L, Wang S, Li T (2017) Md-miR156ab and Md-miR395 target WRKY transcription factors to influence apple resistance to leaf spot disease. Front Plant Sci 8:526. https://doi.org/10.3389/fpls.2017.00526
Zhang Y, Zhang Q, Hao L et al (2019) A novel miRNA negatively regulates resistance to Glomerella leaf spot by suppressing expression of an NBS gene in apple. Hortic Res 6:93. https://doi.org/10.1038/s41438-019-0175-x
Funding
No funding was received.
Author information
Authors and Affiliations
Contributions
JS, CK and HP wrote the first draft of the manuscript. DK and SD wrote some sections of the manuscript and contributed in manuscript editing and revision.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflicts of interest with the present manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kulshrestha, C., Pathak, H., Kumar, D. et al. Elucidating micro RNAs role in different plant–pathogen interactions. Mol Biol Rep 47, 8219–8227 (2020). https://doi.org/10.1007/s11033-020-05810-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05810-y