Abstract
The ica genes in methicillin-resistant Staphylococcus aureus (MRSA) play an important role in biofilm formation. The aim of this study is to define effect of antibiotic resistance and clinical specimens to the expression of ica genes based on their sequence types (STs) and clonal complex (CC). One-hundred (100) S. aureus strain were collected from two teaching therapeutic centers in Hamedan, Iran. Then, the PCR, qPCR, and MLST were used to characterize strains. The results indicated that 29 (29%), 15 (15%), and 5 (5%) strain were strong, mediate, weak biofilm producer, respectively, and the icaA (17%) and icaC (14%) genes were the most abundant. However, two unique STs (3667, 491) in Iran were reported and ST30 and ST11 were the most abundant STs and CC30 and CC5 were observed among MRSA and MSSA strains. High activity in ica locus was observed among strains collected from wound and catheter strains. Also, expression level of icaA gene increased in all strains except ST30 and ST491. Moreover, the highest expression level was observed in CC1, CC7, and CC11. Likewise, activity of the icaC gene was only observed in CC5. Furthermore, the expression of all ica genes in CC5 was significantly correlated with the type of biofilm and the clinical sample. In this study demonstrated that the frequency distribution of STs and CCs in different strains of MRSA was higher than methicillin-sensitive strains. Also, the type of clinical specimen and expression of ica genes played an important role in this abundance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Antimicrobial resistance in Staphylococcus aureus was first identified in the 1940s with the isolation of penicillin-resistant strains [1]. One year after introducing Methicillin- a synthetic derivative of penicillin- in 1959, first strains of methicillin-resistant S. aureus (MRSA) have been emerged [2]. Nowadays, MRSA remains a global health care issue empowered through acquiring resistance against multiple classes of antibiotics [3]. Living in a biofilm gives the bacteria the advantage of a better adaptation to environmental factors and increased resistance to hostile conditions [4]. It is becoming increasingly more difficult to treat bacteria with antimicrobial agents due to an increase in the resistance to antimicrobial compounds which occurs either through the spread of resistance genes, generalized stress response mechanisms, or by the formation of a biofilm [5, 6].
Biofilms can provide protection in a number of different ways. The exopolysaccharide present in the biofilm can act as a physical barrier inhibiting the entry of antimicrobial agents and antibodies into the biofilms [7]. There are two major ways in which biofilm forms, one relies on the ica operon and poly-N-acetyl-β-(1–6)-glucosamine (PNAG) production, while the second is ica independent. The icaADBC encodes four genes including icaA, icaB, icaC, and icaD [8]. icaA and icaD, which collectively produce PIA, facilitate the cells binding together and forming into biofilms. The majority of S.aureus strains contain the icaADBC operon which is upregulated under in vivo conditions [9]. The process of S. aureus biofilm formation is controlled by quorum sensing which is a system used by bacteria for cell–cell communication to regulate gene expression in response to the cell density. The staphylococcal accessory gene regulator (agr) system plays an important role in QS [10], by activating some PIA (polysaccharide intercellular adhesin) dependent surface factors, the system can increase the pathogenicity of S. aureus. In several studies, this system plays the role of downregulation in bacterial colonization and upregulation in host disease [9]. The downregulation and upregulation of the genes involved in the described processes promote the establishment and development of MRSA infections. In addition, these genes play an important role in MRSA biofilm formation which, in turn, leads to a more aggressive infection giving the patient a poor prognosis [11, 12].
Staphylococcus aureus lineages are commonly described by their clonal complex (CC) or sequence type (ST), as determined by molecular typing method. Molecular epidemiology analyses of globally derived S. aureus strain indicate the most prevalent lineages in CC1, CC5, CC8, CC9, CC12, CC15, CC22, CC25, CC30, CC45 and CC51 [1]. Although all of the described lineages are common among MSSA strain, their distribution is more limited among the MRSA clones [13].
Hence, the aim of this study is to define effect of antibiotic resistance and clinical specimens to the expression of ica genes based on their sequence types (STs) and clonal complex (CC). Also, for investigate the relationship between different variables, STs and CCs were identified in different strains of S. aureus based on MLST typing.
Material and methods
Study design and sampling
In this cross-sectional study, 100 strain of S. aureus collected from 492 different clinical samples (including blood, urine, wound infection and catheter) of Hamadan’s Hospitals, Hamadan, Iran between Jun 2017 and Oct 2018 (In this case, the patient was not directly sampled and the strain were collected from the hospital laboratory). Sample size was selected using a simple random sampling technique based on inclusion and exclusion criteria [14]. All strain were transferred to the Microbiology Laboratory of Hamedan University of Medical Sciences for differential tests.
Detection of S. aureus
Conventional identification of S. aureus from human infection is based on microbial culture of clinical samples, including examination of colony morphology and hemolysis type on blood agar after incubation for 18-24 h at 37 °C, Gram stain morphology, catalase reaction, and coagulase reaction. All samples were inoculated onto 5% sheep blood agar base (Merck, Darmstadt, Germany) and mannitol salt agar (Merck, Darmstadt, Germany) and incubated for 18–24 h at 37 °C and finally confirmed as S. aureus on DNase agar (Merck, Darmstadt, Germany) [15]. Based on our observation, some phenotypic tests were optimized based on workflow and laboratory conditions. In this case, we used high salt concentration (10%NaCl) for inhibits the growth of all bacteria except S. aureus. Also, due to contamination of urine samples with Proteus mirabilis, Blood Agar medium with 5% agar was used for urine samples. Finally, ITS gene was used for molecular confirmation of S. aureus strains. All strain were stored in Brain heart infusion broth (Merck, Germany) medium containing 15% glycerol at − 20 °C. This study was approved by the Ethics Committee of Hamadan University of Medical Sciences (Code No: IR.UMSHA.REC.1395.757).
Antimicrobial susceptibility testing and MRSA detection
Antimicrobial susceptibility test was investigated by the Kirby Bauer disc diffusion method according to Clinical Laboratory Standards Institute (CLSI) guidelines. The following drugs and concentrations were used to determine the antibiotic resistance of the strains: penicillin (10U), tetracycline (30 μg), clindamycin (30 μg), gentamicin (30 μg), ciprofloxacin (30 μg), erythromycin (15 μg), rifampin (5 μg) and linezolid (30 μg). All antibiotic disks were obtained from MAST® Company, United Kingdom. MRSA strains was determined using th cefoxitin E-test (Liofilchem, Italy) for Minimum Inhibitory Concentration (MIC) determination of strains. S. aureusATCC25923 and S. aureusATCC43300 were used as the negative and positive control, respectively [16].
Detection of biofilm forming strains in S. aureus
Biofilm formation was determined via crystal violet staining method according to the procedure described by Côrtes et al. [17]. A biofilm unit (BU) was defined as proposed by Côrtes et al. and the strain were classified as non-producers (BU ≤ 0.230) or as weak (0.230 > BU ≤ 0.460), moderate (0.460 > BU ≤ 0.920) or strong producers (BU > 0.920).
Extraction of genomic DNA
DNA was extracted from S. aureus samples using the Qiagen DNA extraction kit (Qiagen, Germany), with modifications. Briefly, strain were sub-cultured onto Tryptose Agar (Merck, Germany) and incubated overnight at 37 °C. Between four to eight colonies were then inoculated into 10 mL sterile plastic tubes containing a 6 mL LB broth (Merck, Germany) and incubated for a further 18–36 h at 37 °C. Cell pellets were prepared from the broth by centrifuging the tubes at 8000 rpm for 10 min. The supernatant was removed except for 50 µL liquid, then the cell pellet and liquid were transferred to 1.5 mL microcentrifuge tubes before being centrifuged a second time at 8000 rpm for 5 min. Cell pellets were then frozen at − 20 °C. To achieve cell wall lysis, frozen cell pellets were resuspended in 180 μL lysozyme 200 mg/mL (Sigma Aldrich, USA) and incubated at 37 °C for 1 h. DNA extraction then continued following the Qiagen DNA kit protocol. DNA yield and purity (A260/A280) were measured using the spectrophotometer Nanodrop (Hangzhou Allsheng Instruments Co., Ltd, Chaina).
Screening of ica genes by PCR method
The ica genes were amplified using specific primers and conditions described in Table 1 [18]. For all genes, the PCR was done in 25 µL reactions containing 1 µL of DNA template, 12 µL of 2.5X master mix (Fermentas, United States), 1 µL of each primer and 10 µL of deionised water. The programmable thermal cycler (Eppendorf, Mastercycler® 5332, Germany) PCR device was applied in all PCR reactions. The PCR conditions were modified as follows: denaturation at 94 °C for 10 min, followed by 25 cycles of denaturation at 94 °C for 45 s, annealing at 57 °C for 45 s and extension at 72 °C for 75 s and final extension step at 72 °C for 10 min. PCR products were separated in 1% agarose gel for 95 min at 85 V, stained with GelRed 3X (Biotium, Fremont, CA) and detected by UV trans illumination. S. aureusATCC25923 and S. aureusATCC43300 were used as the negative and positive control, respectively. Finally, among the collected strain of S. aureus, 16 strain for ica genes with the highest frequency were selected to evaluate the gene expression.
Measurement of ica genes expression levels
The total RNA was extracted using RiboEx kit (GeneAll, Korea) and cDNA synthesized by Hyper ScriptTM Reverse Transcriptase kit (GeneAll, Korea). q-PCR experiments were performed in a total volume of 20 µL including 1 µL of target cDNA, 1 µL of each primer (100 nM), and 5 μL fluorescent EvaGreen dye (Solis BioDyne, Estonia) by a StepOne Plus device (ABI, USA). Amplification process was performed as 95 °C for 15 min followed by 95 °C for 30 s, 59–60 °C (according to the melting temperature of the primers) for 30 s, and 72 °C for 30 s, with data collection in each cycle at 72 °C. Relative expression is used in which the expression of a target gene is standardized by a non-regulated reference gene. All the experiments were performed as triplicate.
MLST typing
All MRSA and MSSA biofilm producing strains were further typed by Multi-Locus Sequence Typing (MLST) based on the study of Strommenger et al. [19]. The amplification products of 7 house-keeping genes including arcC, aroE, glpF, pta, gmk, tpi and yqiL were purified and sequenced in both the directions. Then, the data were analyzed and assigned to sequence types using the tools on the S. aureus MLST webpage.
Statistical analysis
Data were analyzed using Wilcoxon signed rank test, t-test, and Chi-square tests using SPSS version 16(SPSS, Chicago, IL, USA). Statistical significance was defined as the p-value < 0.05.
Results
Prevalence of S. aureus in clinical strain
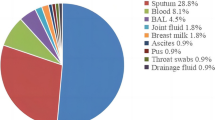
Among 521 different clinical samples, 100 S. aureus were collected including 79 (79%) from female and 21 (21%) strain from male patients. Also, out of 100 clinical strain of S. aureus, 42(42%) strain from wound, 21(21%) strain from blood culture, 24(24%) strain from urine, and 23(23%) strain from catheter were collected (Table 2). The ITS gene was detected in 100 (100%) of the S. aureus strain.
Prevalence of antibiotic resistance and MRSA strains
Figure 1a, b shows that strain had a very high level of antimicrobial resistance. Out of 100 S. aureus, 12 strain (12%) were resistant to linezolid, 17 strain (17%) were resistant to rifampin, 66 strain (66%) were resistant to erythromycin, 12 strain (69%) were resistant to erythromycin, 83 strain (83%) were resistant to tetracycline, 88 strain (88%) were resistant to ciprofloxacin, and 91 strain (91%) were resistant to gentamicin. Also, all strain (100%) resistant were to penicillin and 44(44%) strain were resistant to cefoxitin (MIC ≥ 8 µg/mL) which were considered as MRSA strains. However, 22 MDR strain (22%) and 11 XDR strain (11%) were detected.
Antibiotic resistance and ica gene expression in different clinical strain of S. aureus. a Antibiotic Resistance Pattern in Clinical Strain of S. aureus strains. b Antibiotic Resistance Pattern in Clinical Strain of MRSA strains. c The expression of ica genes in MRSA and MSSA strains and clinical samples. Bars represent means ± SD of the results of three independent experiments. Asterisks indicate significant differences in gene expression levels between (*P < 0.05, **P < 0.01, ***P < 0.001). Dotted horizontal lines represent limit of detection
Prevalence of biofilm producer strains
According to Tables 2 and 3, from the total number of 100 S. aureus strain tested for biofilm formation, strong biofilm producers were 29 (29%), 15 (15%) were moderate and 5 (5%) strain were considered as non or weak biofilm producers. In addition, strong biofilm forming strain were detected in 23 MRSA (52.27%) and 6 MSSA (9.09%) strain.
Prevalence of ica genes
The results of biofilm gene distribution are shown in Tables 2 and 3. Out of 100 strain of S. aureus, 17 strain (17%) were positive for icaA, 9 strain (9%) were positive for icaB, 14 strain (14%) were positive for icaC, 7 strain (7%) were positive for icaD and 11 strain (11%) were positive for icaR.
The frequency of ica genes among 17 strong biofilm producing strains was as following: 15 (88.23%) icaA gene, 7 (41.11%) icaB, 10 (58.88%) icaC isolate, and 5 (29.41%) icaD. In S. aureus with moderate biofilm, 2 strain (13.33%) were positive for icaA gene, 1 (6.66%) for icaB, 2 (13.33%) for icaC, 2 (13.33%) for icaD, and 2 (13.33%) for icaR. And among weak biofilm formers, 1 isolate (20%) possesses icaB, 1 (20%) icaC, and 2 (40%) icaD.
Analysis of MLST typing of S. aureus strain
One-Hundred (100) of S. aureus strain was analyzed and typing by MLST and shown in Fig. 3. However, our analysis revealed a broad phylogenetic distribution of the S. aureus strain included in this study. We detected 32 different multilocus sequence types (ST), two of which (isolate ST3667 and ST491) has not been described before, indeed, each of them detected in single patients. A unique case of S. aureus ST3667 and ST491 harboring all ica genes, except icaR. Further, in S. aureus strains, CC30, ST30, ST5 and ST7 and CC5 had the highest distribution, and ninety of the 100 strain were of ST30 origin. Additionally, we identified 11 and 6 strain, belonging to the broadly distributed MDR high-risk clones ST5 and ST30, respectively. The absence of ST30, ST5 and ST7 in female patients more than male potions. The highest frequency of icaB, iacA and icaD genes was reported in ST30 and ST5 and the highest frequency of icaC genes was observed in ST30 and ST70. Also, the ica genes were most abundant in CC30 and CC5. In addition, high frequency of ST30, ST7 and ST5 was reported in bacteria isolated from wound and blood infection. Prevalence of CC30 and CC5 in bacteria isolated from wound and urinary tract infections was higher than other clinical samples.
Based on Fig. 2, the frequency of STs and CCs in the 44 MRSR strains was as follows: CC30 in 13 strain (29.45%), CC5 in 11 strain (25%), CC7 in 10 strain (22.72%), CC8 in 4 strain (9.09%), CC1 in 3 strain (6.81%) and CC22 in 1 strain (2.27%).
Phylogenetic analysis of multilocus sequence types (STs) of bacteremia S. aureus strain. The neighbor-joining tree was constructed with the concatenated sequences of the seven MLST genes (arcc, aroe, glpf, gmk, pta, tpi, and yqil) based on the distance matrix of pair-wise differences between STs. Blue circle: MSSA; strains; red circle: MRSA strains; and black circle: MDR strains. (Color figure online)
Relative expression report
As illustrated in Figs. 1c, 3, 4, high activity in ica locus was observed among strains collected from wound and catheter isolated strains. Furthermore, it is indicated higher expression level of ica genes in methicillin-resistant strain than methicillin-sensitive strains. Furthermore, the expression of icaA and icaD was high in all biofilm-producing bacteria.
Differences in the expression levels of the ica genes, icaA, icaB, icaC, icaD and icaR, in MRSA and MSSA strain and different Sequence Types. (MSSA strains: ST30, ST22, ST 491) (MRSA strains: ST1, ST5, ST7, ST11, ST30, ST3667). Bars represent means ± SD of the results of three independent experiments. Asterisks indicate significant differences in gene expression levels between (*P < 0.05, **P < 0.01, ***P < 0.001). Dotted horizontal lines represent limit of detection
a Relationship between the expression level of icaA gene, CC, and biofilm formation. b Relationship between the expression level of icaB gene, CC, and biofilm formation. c Relationship between the expression level of icaC gene, CC, and biofilm formation. d Relationship between the expression level of icaD gene, CC, and biofilm formation. e Relationship between the expression level of icaR gene, CC, and biofilm formation. Bars represent means ± SD of the results of three independent experiments. Asterisks indicate significant differences in gene expression levels between (*P < 0.05, **P < 0.01, ***P < 0.001). Dotted horizontal lines represent limit of detection
As displayed in Fig. 3, icaA expression level increased in all investigated strain except for ST30 and ST491. Moreover, the highest expression level was observed in CC1, CC7, and CC11. No activity of icaB gene was observed in the weak biofilm producing strain. The icaC gene was only active in the CC5. Additionally, the activity of all these genes, along with the icaR gene in the CC5 isolate was related to strong, moderate, and weak biofilm with an increasing trend.
Correlation of the ica locus prevalence with MRSA resistance
As shown in Tables 4 and 5, a significant relationship was observed between antibiotic resistance and ica genes. In addition, there is a substantial relationship between the MRSA strains and MSSA strains with biofilm production.
Correlation of the ica locus expression with MRSA resistance and clinical specimen
As illustrated in Fig. 3, a noticeable expression in ica loci was observed in CC5 and CC1. Based on the results of statistical analysis, a significant relationship was detected between the increased expressions of ica genes with methicillin resistance (p < 0.03). In the other words, MRSA strains have a strong biofilm compared to MSSA strains. Moreover, there was a significant relationship between male and female patients and an increase in the expression of ica genes. Also, wound and catheter specimens were associated with a rise in the expression of ica genes (p < 0.08, p < 0.01).
Discussion
Based on Fig. 1a, b in the present study, more than 90% of S. aureus strain were resistant to penicillin and gentamicin. In addition, the resistance to lienozolide and rifampin was found in less than 20% of strain and most strain with antibiotic resistance were wound and blood samples. These results are consistent with the studies of Abubakar and Sulaiman and Khosravi et al. [20, 21]. In our study, the frequency of MRSA and MSSA strains was 44% and 66%, respectively, which are in line with the results of Sit et al. [22]. However, in some studies in the Netherlands [23], Turkey [24], and UK [25], the results were inconsistent with those of the present study with less than 20% frequency in MRSA strains.
Our analysis of Tables 2 and 3 indicated that biofilm forming S. aureus had a high prevalence in MRSA strains. This results agree with Loughman et al. study [26]. Furthermore, the high frequency of MRSA strain with biofilm forming ability in the present study showed that virulence factors are involved in increasing the resistance to methicillin. In line with our findings, Algburi et al. concluded that biofilms play an important role in increasing antibiotic resistance [27]. Jimi et al. reported that there is a significant relationship between methicillin resistance and biofilm formation in S. aureus [28].
Present study described that icaA and icaC genes with 17% and 14% had the highest frequency, while icaD (7%) and icaB (9%) genes had the lowest prevalent. Additionally, based on Tables 2 and 3 nine MDR and XDR strains (2.04%) were found among methicillin-resistant S. aureus strongly produced biofilm. Therefore, no icaR gene was observed in S. aureus with a strong biofilm. Cerca et al. indicated that the icaR gene plays an important role in inhibiting biofilm production and suppressing ica locus, and that icaR fails to play a significant effect on the global regulator of virulence genes such as SarA and agr expression [29]. These results conformed a significant relationship between the frequency of icaA, icaB, icaC, icaD and icaR genes with resistance to methicillin in S. aureus. Further, the extension of these genes was significantly associated with the type of clinical specimen, and the highest frequency of ica locus was observed in the wound and catheter specimens. In order to confirm the results, Serray et al. [30] reported a significant correlation between the frequency of ica genes and antibiotic resistance in S. aureus.
As shown in Fig. 1c, it is found that the expression of ica genes increased in MRSA strains. This figure released that gene expression if ica profiles of S. aureus in a biofilm differ significantly from those clinical sample source, which contributes to antibiotic resistance, evasion of immune responses and expression of virulence factors. Proteins active within a biofilm population undergo up and down regulation as antibiotics are introduced to the environment to counter the action of the antibiotic. Further, studies by Kot et al., [31] and Piechota et al., [32] demonstrated that in biofilm of S. aureus, genes such as ica are actively expressed and can result in β-lactam resistance. Figure 1c also showed that wound and catheter specimens were associated with a rise in the expression of ica genes (p < 0.08, p < 0.01). Despite the importance of S. aureus acquired antibiotic resistance the innate resistance also plays a crucial role in the widespread of antimicrobial resistance. Furthermore, both acquired and innate resistances are closely linked to the increased pathogenesis of device-related biofilm infections [32].
As illustrated in Figs. 3 and 4, high activity in ica locus was observed among strains collected from wound and catheter isolated strains. Furthermore, the relationship between biofilm formation and the expression of ica genes was assessed; This result was in agreement with Piechota et al. [32] study.
As displayed in Fig. 3, icaA expression level increased in all investigated strain except for ST30 and ST491. Moreover, the highest expression level was observed in CC1, CC7, and CC11. No activity of icaB gene was observed in the weak biofilm producing strain and icaC gene was only active in the CC5. This is consistence with observation of Tasse et al. [33] study.
In Figs. 2 and 3, we indicated that strong biofilm producing strains of MRSA group abundantly belonged to CC5, CC30, CC22, and CC7. The first reported CC491 and CC3667 were observed among weak and moderate biofilm producers with the lowest frequency in MRSA strains and MSSA. Vanhommerig et al. [34] reported that CC5 and CC22 are more abundant in MRSA strains with strong biofilms. Furthermore, according to Table 5 and Fig. 4 a significant relationship was observed between CCs and resistance to methicillin in S. aureus. In another study, Challagundla et al. [35] reported that CC5 and CC22 showed high pathogenicity in MRSA strains and the frequency of antibiotic resistance is higher in these CCs. Additionally, in Figs. 3 and 4, CC30 was observed in non-biofilm forming strains, medium biofilm producers, and strong biofilm forming strains. However, some studies in Ireland [9], Brazil [13], and United Kingdom [36] have shown that the horizontal transfer of virulence and resistance genes between similar CCs and STs might play no role in pathogenicity.
In conclusion, Biofilm formation is different in methicillin-sensitive and methicillin-resistant S. aureus which appear in weak, moderate, and strong states. Based on our findings, the formation of biofilms in MRSA strains is much more than MSSA strains, and the frequency of ica genes can vary in different antibiotic patterns. Furthermore, the diversity of CCs and STs in strains with moderate and strong biofilms indicates extensive genetic movements in drug-resistant strains and plays an important role in virulence and resistance to antibiotics. Hence, it is concluded that resistance to methicillin is affected by pathogenicity due to the significant relationship between the presence of ica genes and antibiotic resistance patterns. Finally, regarding the high expression of the studied genes, biofilm formation in MRSA strains was more dependent on ica locus.
References
Heydari N, Alikhani MY, Jalilian FA et al (2017) Evaluation of real time PCR for detection of clinical isolates of Staphylococcus aureus and methicillin-resistance strains based on melting curve analysis method. Koomesh 19(4):877–886
Pasandideh NK, Habibi MR, Tahmasebi H et al (2018) Activity of biofilm genes icaA and icaR in methicillin-resistant Staphylococcus aureus treated with vitamin K in wound specimens. Koomesh 20(3):588–593
Boswihi SS, Udo EE, Al-Sweih N (2016) Shifts in the clonal distribution of methicillin-resistant Staphylococcus aureus in Kuwait hospitals: 1992–2010. PLoS ONE 11(9):e0162744
Udo EE, Boswihi SS (2017) Antibiotic resistance trends in methicillin-resistant Staphylococcus aureus Isolated in Kuwait Hospitals: 2011–2015. Med Princ Pract 26(5):485–490
Vafaeefar M, Yousef Alikhani M, Tahmasebi H et al (2017) Identification and determination of the relationship between ccr alleles and antibiotic resistance in clinical isolates of methicillin resistant Staphylococcus aureus. J Babol Univ Med Sci 19(12):28–35
Bongiorno D, Mongelli G, Stefani S et al (2017) Burden of rifampicin- and methicillin-resistant Staphylococcus aureus in Italy. Microbial Drug Resist 24(6):732–738
Sritharadol R, Hamada M, Kimura S et al. (2018). Mupirocin at subinhibitory concentrations induces biofilm formation in Staphylococcus aureus. Microb Drug Resist.
Ghasemian A, Najar-Peerayeh S, Bakhshi B et al (2015) High prevalence of icaABCD genes responsible for biofilm formation in clinical isolates of Staphylococcus aureus from hospitalized children. Arch Pediatr Infect Dis 3(3):e20703
McCarthy H, Rudkin JK, Black NS et al (2015) Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front Cell Infect Microbiol 5:1–1
Dehbashi S, Tahmasebi H, Zeyni B et al (2018) The relationship between promoter-dependent quorum sensing induced genes and methicillin resistance in clinical strains of Staphylococcus aureus. J Zanjan Univ Med Sci 26(116):75–87
da Fonseca W, Batistão D, Amaral de Campos P, Caroline Camilo N et al (2016) Biofilm formation of Brazilian meticillin-resistant Staphylococcus aureus strains: prevalence of biofilm determinants and clonal profiles. J Med. Microbiol 65(4):286–297
Long SW, Olsen RJ, Mehta SC et al (2014) PBP2a Mutations causing high-level ceftaroline resistance in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrob Agents Chemother 58(11):6668–6674
Teixeira MM, Araújo MC, Silva-Carvalho MC et al (2012) Emergence of clonal complex 5 (CC5) methicillin-resistant Staphylococcus aureus (MRSA) isolates susceptible to trimethoprim-sulfamethoxazole in a Brazilian hospital. Br J Med Biol Res 45(7):637–643
setia m s. (2016) Methodology series module 5: sampling strategies. Indian J Pediatr 61(5):505–509
Bokaeian M, Tahmasebi H (2017) Molecular identification of genes responsible for resistance to aminoglycosides and methicillin in clinical samples of Staphylococcus aureus. J Babol Univ Med Sci 19(3):38–46. https://doi.org/10.22088/jbums.19.3.38
Clinical and Laboratory Standards Institute (2017) Performance standards for antimicrobial susceptibility testing. twenty-seventh informational supplement. M100S, 27 edn. CLSI, Wayne
Côrtes MF, Beltrame CO, Ramundo MS et al (2015) The influence of different factors including fnbA and mecA expression on biofilm formed by MRSA clinical isolates with different genetic backgrounds. Int J Med Microbiol 305(1):140–147
Solati SM, Tajbakhsh E, Khamesipour F et al (2015) Prevalence of virulence genes of biofilm producing strains of Staphylococcus epidermidis isolated from clinical samples in Iran. AMB Express 5(1):47
Strommenger B, Kettlitz C, Weniger T et al (2006) Assignment of staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J Clin Microbiol 44(7):2533–2540
Abubakar U, Sulaiman SAS (2018) Prevalence, trend and antimicrobial susceptibility of methicillin resistant Staphylococcus aureus in Nigeria: a systematic review. Int J Infect Control 11(6):763–770
Khosravi AD, Jenabi A, Montazeri EA (2017) Distribution of genes encoding resistance to aminoglycoside modifying enzymes in methicillin-resistant Staphylococcus aureus (MRSA) strains. Kaohsiung J Med Sci 33(12):587–593
Sit PS, Teh CSJ, Idris N et al (2017) Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) infection and the molecular characteristics of MRSA bacteraemia over a two-year period in a tertiary teaching hospital in Malaysia. BMC Infect Dis 17(1):274–274
Ravensbergen S J, Berends M, Stienstra Y et al. (2017). High prevalence of MRSA and ESBL among asylum seekers in the Netherlands. PLoS ONE, 12(4), e0176481.
Rağbetli C, Parlak M, Bayram Y et al (2016) Evaluation of antimicrobial resistance in Staphylococcus aureus isolates by years. Jpn J Infect Dis 2016:9171395–9171395
Horner C, Wilcox M, Barr B et al. (2012). The longitudinal prevalence of MRSA in care home residents and the effectiveness of improving infection prevention knowledge and practice on colonisation using a stepped wedge study design. BMJ Open, 2(1).
Loughman JA, Fritz SA, Storch GA et al (2009) Virulence gene expression in human community-acquired Staphylococcus aureus infection. Int J Infect Dis 199(3):294–301
Algburi A, Comito N, Kashtanov D et al. (2017). Control of biofilm formation: antibiotics and beyond. Appl Environ Microbiol, 83(3).
Jimi S, Miyazaki M, Takata T et al (2017) Increased drug resistance of meticillin-resistant Staphylococcus aureus biofilms formed on a mouse dermal chip model. J Med Microbiol 66(4):542–550
Cerca N, Brooks JL, Jefferson KK (2008) Regulation of the intercellular adhesin locus regulator (icaR) by SarA, sigmaB, and IcaR in Staphylococcus aureus. J bacteriol 190(19):6530–6533
Serray B, Oufrid S, Hannaoui I et al (2016) Genes encoding adhesion factors and biofilm formation in methicillin-resistant Staphylococcus aureus in Morocco. J Infect Dev Ctries 10(8):863–869
Kot B, Sytykiewicz H, and Sprawka I. (2018). Expression of the Biofilm-Associated Genes in Methicillin-Resistant Staphylococcus aureus in Biofilm and Planktonic Conditions. Int J Mol Sci, 19(11).
Piechota M, Kot B, Maciejewska AF et al (2018) Biofilm formation by methicillin-resistant and methicillin-sensitive Staphylococcus aureus strains from hospitalized patients in Poland. Biomed Res Int 2018:7
Tasse J, Trouillet-Assant S, Josse J et al (2018) Association between biofilm formation phenotype and clonal lineage in Staphylococcus aureus strains from bone and joint infections. PLoS ONE 13(8):e0200064–e0200064
Vanhommerig E, Moons P, Pirici D et al (2014) Comparison of biofilm formation between major clonal lineages of methicillin resistant Staphylococcus aureus. PLoS ONE 9(8):e104561–e104561
Challagundla L, Reyes J, Rafiqullah I et al (2018) Phylogenomic classification and the evolution of clonal complex 5 methicillin-resistant Staphylococcus aureus in the western hemisphere. Front Microbiol 9:1901–1901
Marbach H, Boakes E, Lynham S et al (2017) Identification of a distinctive phenotype for endocarditis-associated clonal complex 22 MRSA isolates with reduced vancomycin susceptibility. J Med Microbiol 66(5):584–591
Acknowledgements
The authors of this article are grateful to Hamadan University of Medical Sciences for their financial support in conducting research.
Author information
Authors and Affiliations
Contributions
MRZ designed research; HT, SD and MJ performed experiments. HT and MRA wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tahmasebi, H., Dehbashi, S., Jahantigh, M. et al. Relationship between biofilm gene expression with antimicrobial resistance pattern and clinical specimen type based on sequence types (STs) of methicillin-resistant S. aureus. Mol Biol Rep 47, 1309–1320 (2020). https://doi.org/10.1007/s11033-019-05233-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-05233-4