Abstract
The combination of gemcitabine and cisplatin has been shown previously to elicit a synergistic therapeutic effect on bladder cancer cell lines and result in reduced cell survival. However, the precise mechanism by which cells die has not been elucidated. Cell cycle-related genes are the predominant targets of chemotherapeutic protocols. Therefore, molecular biomarkers that are predictive of therapeutic outcomes associated with tumor sensitivity might be important for optimal treatment protocol selection. The aim of this study was to investigate the changes in gene expression in cell cycle-related genes that were induced by cisplatin, gemcitabine or a combined treatment using both agents in a low-grade urinary bladder transitional carcinoma cell line (RT4). The following three treatment protocols were used: 1.0 μM cisplatin, 1.56 μM gemcitabine and a combination of 1.0 μM cisplatin and 1.56 μM gemcitabine. Cytometry and morphology analysis (by phase-contrast photomicrography) were performed in addition to pathway-specific gene expression analysis using quantitative RT-PCR gene arrays. The following results were observed after 1.0 μM cisplatin treatment: (1) a decrease in cell number, (2) an increased percentage of scattered cells and (3) downregulated expression of genes related to cell cycle arrest, G1/S-to-mitotic cell cycle transition, DNA repair, apoptosis, transcription and mitosis. Treatment with 1.56 μM gemcitabine, or with both drugs simultaneously, induced the following effects: (1) a decrease in cell number, (2) an increased percentage of scattered and elongated cells, (3) the modulation of genes that are predominantly involved in DNA repair and (4) a significant upregulation of genes related to cell cycle arrest. Reduced cell density was observed after the combined treatment compared to the two other single-agent protocols. The downregulation of MRE11A and SKP2 was observed only in cells subjected to the combined treatment. In conclusion, cisplatin, gemcitabine and the combination of both drugs elicited distinct toxicogenomic effects in the RT4 bladder transitional carcinoma cell line, although disruptions in the expression of cell cycle control-related genes and other pathways responsible for cell survival were observed for all of the protocols. MRE11A and SKP2 downregulation appeared to be responsible for the synergistic therapeutic effects elicited by cisplatin and gemcitabine.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cancer is a serious public health problem. Bladder cancer is one of the costliest tumor types for health care systems to treat, particularly because of the need for routine clinical monitoring (urine cytology and cystoscopy). Bladder cancer is predominantly of the transitional cell type (TCC), and 90 % of all bladder carcinomas are classified as this type [1]. Approximately 70 % of all TCC tumors are confined to the epithelium or subepithelial connective tissues, and the remaining 30 % exhibit solid and invasive growth patterns [2]. Patients with non-muscle-invasive urothelial tumors experience high rates of recurrence and progression despite aggressive therapy [3]. A previous study conducted in our laboratory revealed that genetic instabilities remain in urothelial cells from patients after tumor resection and chemotherapy [4].
To standardize chemotherapeutical protocols, the genes associated with tumor resistance or sensitivity to antineoplastic drugs must be identified. The resulting molecular biomarkers that are predictive of therapy outcomes might help clinicians to choose the most appropriate treatments. Regarding bladder cancer, some studies have shown that gemcitabine/cisplatin regimens elicit a similar efficacy as the methotrexate, vinblastine, doxorubicin and cisplatin (MVAC) protocol but with superior safety and tolerability [5]. These two drugs exhibit different mechanisms of action. Whereas cisplatin induces DNA cross-linking and causes severe lesions that can lead to apoptosis [6], gemcitabine is a deoxycytidine analog, which becomes an active dFdCTP metabolite when phosphorylated, and is incorporated into DNA, thereby blocking replication [7].
High-throughput methods can help to elucidate molecular mechanisms in different cell types, promoting a more comprehensive understanding of carcinogenesis, tumor development and chemotherapeutic protocols [8]. Using DNA microarray technology, some studies have shown that the repression of the hTERT (human telomerase reverse transcriptase) transcript decreases the expression of the EGFR gene, suggesting that hTERT promotes tumor growth by mechanisms that are independent of telomere lengthening [9]. Using the same technology, Missiaglia et al. [10] have associated the increased expression of CDKN1A and GADD45A in human pancreatic cancer cells with the therapeutic efficacy of the methylase inhibitor 5-aza-2′-deoxycytidine (5-aza-CdR). Using PCR arrays, other studies have reported the downregulation of the EGF and IGF genes in human prostate cell lines after treatment with genistein or daidzein, demonstrating a chemopreventive activity of isoflavones [11]. In bladder cancer, this methodology was important for showing the possible relationship between the gemcitabine-induced upregulation of the CCNE1, CDKN1A and GADD45A genes in TP53-mutated cells and the cell cycle arrest and modulation of genes related to DNA repair, the G1/S transition and apoptosis. These findings reinforce that the identification of gemcitabine-responsive genes can be used in a clinical setting to predict chemotherapeutic responses [12]. Using cDNA microarrays, Tsunoda et al. [13] have also suggested that the cisplatin-induced downregulation of IP3R1 expression was closely associated with the acquisition of cisplatin resistance in bladder cancer cells. Furthermore, Takata et al. [14] used the same methodology and identified dozens of genes that were differentially expressed between responder and nonresponder tumors in MVAC (methotrexate, vinblastine, doxorubicin and cisplatin) therapy. These authors suggest that the sensitivity of an invasive bladder cancer to the MVAC neoadjuvant chemotherapy can be predicted by gene expression patterns, a step toward the achievement of “personalized therapy”.
Based on these findings, the need to predict therapeutic efficacy and the widespread combined use of cisplatin and gemcitabine, we sought to investigate the effects of both drugs on the expression profile of cell cycle control-related genes in a bladder cancer cell line (RT4). Considering that 70 % of all bladder cancers are confined to the epithelium or subepithelial connective tissues, we used RT4 cells because these cells were originally derived from a superficial, low-grade tumor. The elucidation of the antineoplastic activity of cisplatin and gemcitabine might improve treatment protocols and contribute to the selection of the best medical treatments.
Materials and methods
Cell lines and test compounds
The RT4 cell line (TP53 wild type), derived from a low-grade papillary bladder tumor, was purchased from the Cell Bank of the Federal University of Rio de Janeiro, Brazil. The cells were maintained as previously described by da Silva et al. [15]. The antineoplastic drugs gemcitabine (dFdC, Gemzar) and cisplatin were purchased from Eli Lilly Laboratory (USA) and Sigma-Aldrich, respectively. Ultra-pure sterilized water was used for dilution.
Experimental design
Cells were seeded into 12-well culture plates (5 × 104 cells/well) and evaluated for cell viability, cell count and morphological changes. Cells were seeded into 25 cm3 culture flasks (2 × 106 cells/flask) for RNA extraction. Twenty-four hours after seeding (T1 = 0 h), the cells were treated with gemcitabine (0.78, 1.56 or 3.12 μM), cisplatin (0.5, 1.0 or 2.5 μM) or both drugs simultaneously during a 24-h period (T2 = 24 h). The drug concentrations were defined in previous experiments [15]. Untreated cells were cultured in parallel and constituted a negative control. At T2, the cells were washed with Hank’s solution (0.4 g KCl, 0.06 g KH2PO4, 0.04 g Na2HPO4, 0.35 g NaHCO3, 1 g glucose and 8 g NaCl in 1,000 ml H2O), harvested and evaluated for cell viability and gene expression. Fresh medium was then added, and after another 48-h period (T3 = 72 h), the cells were collected, counted and evaluated for morphological changes.
Cytometry and morphology
Viable cells were counted using a hemocytometer before (20,000 cells) and after the treatments detailed above. Cell viability was evaluated using the trypan blue exclusion test. The assays were performed in triplicate. The morphological changes were analyzed using a phase-contrast microscope before and after gemcitabine and/or cisplatin treatments.
RNA extraction
Total RNA was extracted using the Mini RNeasy kit (Qiagen), according to the manufacturer’s protocols. The extracted RNA was stored at −80 °C. The RNA integrity and quality were evaluated using 2 % denaturing agarose gels and NanoVue equipment, respectively.
PCR arrays
To evaluate changes in the gene expression profile, gemcitabine and cisplatin were used at concentrations of 1.56 and 1.0 μM, respectively. These two concentrations were genotoxic, as evidenced by a comet assay [16], but were not cytotoxic in the trypan blue and XTT tests [15]. A cell cycle pathway PCR array (PAHS-020A—SA Biosciences) was used for the qRT-PCR analysis. The RNA was reverse transcribed using the RT2 First Strand kit (SA Biosciences), according to the manufacturer’s protocol. An aliquot of the diluted first-strand synthesis reaction was added to the SYBR Green/ROX master mix (SA Biosciences) along with nuclease-free water in accordance with the PCR array system’s user manual. Afterward, 25 μl of the cDNA/master mix cocktail was added into each well of the pathway-specific qRT-PCR microplate. The quality controls for genomic DNA contamination, reverse transcription efficiency and PCR amplification efficiency were analyzed. The qRT-PCR array data were normalized using the arithmetic mean values of three housekeeping genes (B2 M, HPRT1 and ACTB). All of the arrays were performed in triplicate. Information about the biological functions of the genes was obtained from FATIGO (http://babelomics.bioinfo.cipf.es/).
Statistical analysis
For the statistical analysis of cell viability and cell number, a one-way ANOVA test was used. For the gene expression analysis, fold change was evaluated. In this case, p values were calculated using Student’s t test on the expression values that were collected in triplicate for each gene in the control and treatment groups. A p value < 0.05 was considered statistically significant.
Results
Table 1 shows the number of cells after treatment with the different antineoplastic protocols. All concentrations of gemcitabine alone, the two highest concentrations of cisplatin alone and all concentrations of the combined drug treatment (i.e., all three treatment protocols) resulted in statistically significant reductions in cell number (p < 0.05). Moreover, statistically significant differences were observed when the combination of 1.0 μM cisplatin and 1.56 μM gemcitabine was compared to treatments with only 1.0 μM cisplatin or 1.56 μM gemcitabine. Statistically significant differences were also observed when comparing the results of the combined treatment with 2.5 μM cisplatin and 3.12 μM gemcitabine to the results of the treatment with either 2.5 μM cisplatin or 3.12 μM gemcitabine.
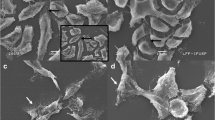
Phase contrast microscopy revealed scattered cells after the cisplatin treatment, while scattered and elongated cells were observed after the gemcitabine and combined treatments. The gemcitabine and combined treatments also led to a reduction in cell number (Fig. 1).
Photomicrography of the human bladder transitional carcinoma cell line RT4 before and after treatments as follows: a control (without treatment), b 1.0 μM cisplatin, c 1.56 μM gemcitabine and d 1.0 μM cisplatin + 1.56 μM gemcitabine. The arrows indicate elongated cells. The scattered cells and cell groups are shown in panels B, C and D. Phase-contrast microscopy, 400×
At the time of sampling for gene expression analyses, the cell viability was always greater than 90 %. Compared to the control treatment, 11/84 (13.1 %), 32/84 (38.1 %) and 31/84 (36.9 %) of the genes were differentially expressed (p < 0.05) after the cisplatin, gemcitabine and combined treatments, respectively (Figs. 2, 3 and 4). The most significantly upregulated genes (fold change >2, p < 0.05) after the gemcitabine treatments were BAX, CCND2, CDC34, CDKN1A, GADD45A, RBBP8 and SERTAD1, while the most significantly downregulated genes were ATM, CCNB1 and CDC20. After the combined treatment with cisplatin and gemcitabine, BAX, CCND2, CDC34, CDK6, CDKN1A, GADD45A, RBBP8 and SERTAD1 were upregulated, while CDC20, MRE11A and SKP2 were downregulated. The CCNB1, CCND2, CDC20, CDKN1A, CDKN3, CUL 2, GADD45A, KPNA2 and SERTAD1 genes were equivalently modulated (upregulated or downregulated) after all of the treatment protocols used (p < 0.05). The significant upregulation of CKS2 was observed exclusively during cisplatin treatment, while the ATR, BCCIP, CCNG1, CDK2, MNAT1, RAD9A and RB1 genes were exclusively modulated after the gemcitabine treatments. The MRE11A and SKP2 genes were exclusively downregulated after the combined treatment (p < 0.05) (Tables 2, 3 and 4).
Gene expression profiling of the human bladder transitional carcinoma cell line RT4 after treatment with 1.0 μM cisplatin. A heat map showing genes that were significantly modulated in three independent replicates. The red spots indicate upregulation, and the green spots indicate downregulation. The black spots indicate the absence of modulation, whereas the gray spots indicate the absence of values. (Color figure online)
Gene expression profiling of the human bladder transitional carcinoma cell line RT4 after treatment with 1.56 μM gemcitabine. A heat map showing the genes that were significantly modulated in three independent replicates. The red spots indicate upregulation, and the green spots indicate downregulation. The black spots indicate the absence of modulation, whereas the gray spots indicate the absence of values. (Color figure online)
Gene expression profiling of the human bladder transitional carcinoma cell line RT4 after the combined treatment (1.0 μM cisplatin + 1.56 μM gemcitabine). A heat map showing genes that were significantly modulated in three independent replicates. The red spots indicate upregulation, and the green spots indicate downregulation. The black spots indicate the absence of modulation, whereas the gray spots indicate the absence of values. (Color figure online)
Discussion
The identification of genes associated with tumor sensitivity to antineoplastic drugs may be potentially important for the selection of more effective chemotherapeutical protocols. Therefore, using bladder transitional carcinoma cells, we investigated whether the effectiveness of different cisplatin and gemcitabine treatment protocols was associated with the expression of a panel of cell cycle-related genes. Prior to analyzing the gene expression profiles, we assessed the effects of gemcitabine and cisplatin on cell proliferation and morphology. We observed reduced cell density after the simultaneous treatment with cisplatin and gemcitabine. This finding is consistent with our previous study, which demonstrated that the combined treatment with gemcitabine and cisplatin induces a G1-phase arrest and triggers apoptosis [15]. The observed morphological changes (scattered and elongated cells) were also suggestive of abrogated cell cycle dynamics. Gonçalves et al. [17] have suggested that irregular morphologies may indicate the possible involvement of the actin cytoskeleton. Additionally, morphological changes and low cell density after gemcitabine treatment were previously reported to be suggestive of cell cycle arrest and apoptosis [15].
For gene expression profiling, the applied concentrations of 1.56 μM gemcitabine and 1.0 μM cisplatin were carefully selected based on our previous findings and showed significant genotoxicity in comet assays [16]; however, these concentrations resulted in low levels of detectable apoptosis (assayed by flow cytometry) [15]. Therefore, based on these earlier observations, our transcriptome profiles were not reflective of dead cells.
Our gene expression arrays showed that CCNB1, CCND2, CDC20, CDKN1A, CDKN3, CUL 2, GADD45A, KPNA2 and SERTAD1 were equally modulated (up- or downregulated) by the three treatment protocols (cisplatin, gemcitabine and cisplatin + gemcitabine). Based on gene ontological analyses, these genes are primarily involved in the negative regulation of the cell cycle (CUL2, CDKN1A, CDKN3 and GADD45A), cell cycle arrest (CDC20, CDKN1A and GADD45A), G1/S transition (CCNB1, CCND2 and CDKN1A), DNA repair (GADD45A), apoptosis (GADD45A), gene transcription (SERTAD1) and mitosis (KPNA2). Several of these genes have been suggested as important targets for cancer treatment. For example, the expression of GADD45A was shown to be associated with the efficacy of 5-aza-CdR in pancreatic cancer cell lines [10], and the upregulation of CDKN1A produced similar effects to those of soy isoflavones on LNCaP (androgen-sensitive human prostate adenocarcinoma) cells [11]. The activation of TP53 has been demonstrated to increase the level of CDKN1A mRNA, which leads to cell cycle arrest at the G1/S transition [18]. Despite the observed upregulation of CDKN1A, we did not detect any modulation of TP53 after the different treatment protocols. This finding was also observed in TP53-mutated bladder transitional carcinoma cells [12], suggesting that both cisplatin and gemcitabine can induce alternative mechanisms of activating CDKN1A that are independent of the TP53 status. The upregulation of CDKN1A induced by the treatment of cells with either cisplatin or gemcitabine might be attributed to cell cycle alterations and arrest, as previously observed in bladder tumor cells [15]. In contrast, the GADD45A upregulation could be related to the decreased cell number observed after the different treatment protocols. We have previously reported that cisplatin, gemcitabine and combined cisplatin and gemcitabine are able to induce apoptosis, but at different times. Herein, gene expression was analyzed only 24 h after treatment. Therefore, genes other than GADD45A might be responsible for the apoptosis rates observed at the different times in our previous study [15]. However, the simultaneous modulation of CDKN1A and GADD45A, which was previously demonstrated in other cell lines [10], might also occur in bladder cancer cells and might be associated with the effectiveness of different treatments.
The CKS2 gene (a negative regulator of the cell cycle) has been proposed to be a potential biomarker for predicting the progression of superficial bladder cancer to muscle-invasive cancer [19]. Herein, we detected the downregulation of CKS2 only after cisplatin treatment. This finding might explain the previously described differences in the cell cycle effects elicited by cisplatin compared to gemcitabine treatments [15]. Whereas most of the cells were in the S phase after treatment with cisplatin, gemcitabine induced a cell cycle arrest at G1. In contrast, ATR (DNA repair), BCCIP (regulation of cell cycle arrest), CCNG1 (negative regulation of the cell cycle), CDK2 (negative regulation of the cell cycle), MNAT1 (G1/S transition of the mitotic cell cycle), RAD9A (negative regulation of the cell cycle) and RB1 (negative regulation of the cell cycle) were only modulated after treating the cells with gemcitabine. Some of these genes have also been demonstrated to be important biomarkers. Recently, Weis et al. [20] reported that the modulation of RAD9A in skin fibroblasts prevented the formation of secondary cancers. Other studies have shown that the ATR gene codes for the kinase that activates DNA repair in Fanconi anemia [21]. Based on our current results, the gemcitabine-induced modulation of genes related to DNA repair (ATR, ATM and RAD9), the cell cycle (CDKN1A and CDK2) and apoptosis (BAX and GADD45A) might explain the G1 arrest and apoptosis rates reported in our previous publication [15].
The majority of the genes that were modulated after the combined treatment (cisplatin + gemcitabine) were the same as those modulated after treatment with gemcitabine alone. However, MRE11A (which is involved in the response to a DNA damage stimulus and the cellular response to stress) and SKP2 (which is involved in the negative regulation of the cell cycle), both of which are tightly linked to cancer, were exclusively downregulated after the combined treatment. Based on the concept of synthetic lethality, studies have shown that MRE11-deficient cells are more sensitive to poly (ADP-ribose) polymerase (PARP-1) inhibition [22]. When DNA is moderately damaged, PARP-1 is activated and participates in the DNA repair process, thus contributing to cell survival. In contrast, during extensive DNA damage, PARP-1 is over-activated and induces a depletion of cellular NAD + and ATP levels, which leads to cellular dysfunction and even necrotic cell death [23]. However, PARP-1 inhibition in cells exposed to DNA-damaging drugs might decrease DNA repair and induce apoptotic cell death but prevent necrosis and other pathological side effects [24]. In fact, we have previously reported increased DNA damage, as detected by the comet assay, and high apoptosis rates, as analyzed by flow cytometry, in RT4 cells after treatment with cisplatin and gemcitabine [15, 16]. The SKP2 protein has been reported to regulate cell proliferation by targeting several cell cycle-regulated proteins for ubiquitination and degradation [25]. Moreover, a recent study has shown that SKP2 overexpression restores the colony formation capacity of cells treated with prodigiosin (a bacterial tripyrrole pigment with strong proapoptotic activity) [26]. Therefore, the increased cytotoxic effects observed after the combined treatment might be explained by the downregulation of MRE11A and SKP2. These two proteins might act by decreasing DNA repair and inducing apoptotic cell death. Alternative mechanisms, such as the inhibition and ubiquitination of PARP-1 as well as the degradation of cell cycle-regulated proteins, might also explain the observed synergistic effects of gemcitabine and cisplatin.
In conclusion, we have observed that the treatment of cells with cisplatin and gemcitabine modulates several molecular pathways, including those governing the G1/S transition, apoptosis and the negative regulation of the cell cycle. In our view, the genes that we have highlighted represent those that would be associated with the efficacy of antitumor therapies. Furthermore, the increased efficacy and synergistic effects of the combined cisplatin and gemcitabine treatment might be attributed to the downregulation of MRE11A and SKP2. The identification of chemotherapeutic-responsive genes provides insight into anticancer mechanisms and might be important in clinical settings to predict chemotherapeutic responses.
References
Cordon-Cardo C (2008) Molecular alterations associated with bladder cancer initiation and progression. Scand J Urol Nephrol 218:154–165
Cheng L, Zhang S, MacLennan GT, Lopez-Beltran A, Montironi R (2011) Bladder cancer: translating molecular genetic insights into clinical practice. Hum Pathol 42:455–481
Rink M, Cha EK, Green D, Hansen J, Robinson BD, Lotan Y, Sagalowsky AL, Chun FK, Karakiewicz PI, Fish M, Scherr DS, Shariat SF (2012) Biomolecular predictors of urothelial cancer behavior and treatment outcomes. Curr Urol Rep 13:122–135
Gontijo AMMC, Marcondes JPC, Elias FN, de Oliveira ML, de Lima RO, Salvadori DM, de Camargo JL (2002) DNA Damage in cytologically normal urothelial cells of patients with a history of urothelial cell carcinoma. Environ Mol Mutagen 40:190–199
Belmut J, Albiol S, Ramirez de Olano A, Pujadas J, Maroto P, On behalf the Spanish Oncology Genitourinary Group (SOGUG) (2006) Gemcitabine in the treatment of advanced transitional cell carcinoma of the urothelium. Ann Oncol 17:113–117
Wang D, Lippard SJ (2005) Cellular processing of platinum anticancer drugs. Nat Rev 4:307–319
Toschi L, Finocchiaro G, Gioia V (2005) Role of gemcitabine in cancer therapy. Future Oncol 1:7–17
Coppée JY (2008) Do DNA microarrays have their future behind them? Microbes Infect 10:1067–1071
Kraemer K, Schmidt U, Fuessel S, Herr A, Wirth MP, Meye A (2006) Microarray analyses in bladder cancer cells: inhibition of hTERT expression down-regulates EGFR. Int J Cancer 119:1276–1284
Missiaglia E, Donadelli M, Palmieri M, Crnogorac-Jurcevic T, Scarpa A, Lemoine NR (2005) Growth delay of human pancreatic cancer cells by methylase inhibitor 5-aza-2′-deoxycytidine treatment is associated with activation of the interferon signalling pathway. Oncogene 24:199–211
Rabiaua N, Kossaïa M, Braud M, Chalabi N, Satih S, Bignon YJ, Bernard-Gallon DJ (2010) Genistein and daidzein act on a panel of genes implicated in cell cycle and angiogenesis by polymerase chain reaction arrays in human prostate cancer cell lines. Cancer Epidemiol 34:200–206
da Silva GN, de Camargo EA, Salvadori DM (2012) Toxicogenomic activity of gemcitabine in two TP53-mutated bladder cancer cell lines: special focus on cell cycle-related genes. Mol Biol Rep 39:10373–10382
Tsunoda T, Koga H, Yokomizo A, Tatsugami K, Eto M, Inokuchi J, Hirata A, Masuda K, Okumura K, Naito S (2005) Inositol 1,4,5-trisphosphate (IP3) receptor type1 (IP3R1) modulates the acquisition of cisplatin resistance in bladder cancer cell lines. Oncogene 24:1396–1402
Takata R, Katagiri T, Kanehira M, Tsunoda T, Shuin T, Miki T, Namiki M, Kohri K, Matsushita Y, Fujioka T, Nakamura Y (2005) Predicting response to methotrexate, vinblastine, doxorubicin, and cisplatin neoadjuvant chemotherapy for bladder cancers through genome-wide gene expression profiling. Clin Cancer Res 11:2625–2636
Da Silva GN, Marcondes JPC, Camargo EA, Sakamoto-Hojo ET, Passos GA, Salvadori DMF (2010) Cell cycle arrest and apoptosis in TP53 subtypes of bladder carcinoma cell lines treated with cisplatin and gemcitabine. Exp Med Biol 235:814–824
Camargo EA, da Silva GN, Gobette CP, Marcondes JP, Salvadori DM (2013) No relationship between the amount of DNA damage and the level of hMLH1 and RASSF1A gene expression in bladder cancer cells treated with cisplatin and gemcitabine. Asian Pac J Cancer Prev 14:5941–5948
Gonçalves EM, Ventura CA, Yano T, Macedo MLR, Generi SC (2006) Morphological and growth alterations in vero cells transformed by cisplatin. Cell Biol Int 30:485–494
Sun T, Yang W, Liu J, Shen P (2011) Modeling the basal dynamics of p53 system. PLoS One 6:e27882
Chen R, Feng C, Xu Y (2011) Cyclin-dependent kinase-associated protein Cks2 is associated with bladder cancer progression. J Int Med Res 39:533–540
Weis E, Schoen H, Victor A, Spix C, Ludwiq M, Scheneider-Raetzke B, Kohlschmidt N, Bartsch O, Gerhold-Ay A, Boehm N, Grus F, Haaf T, Galetzka D (2011) Reduced mRNA and protein expression of the genomic caretaker RAD9A in primary fibroblasts of individuals with childhood and independent second cancer. PLoS One 6:e25750
Shigechi T, Tomida J, Sato K, Kobayashi M, Eykelenboom JK, Pessina F, Zhang Y, Uchida E, Ishiai M, Lowndes NF, Yamamoto K, Kurumizaka H, Maehara Y, Takata M (2012) ATR-ATRIP kinase complex triggers activation of the fanconi anemia DNA repair pathway. Cancer Res 72:1149–1156
Vilar E, Bartnik CM, Stenzel SL, Raskin L, Ahn J, Moreno V, Mukheriee B, Iniesta MD, Morgan MA, Rennert G, Gruber SB (2011) MRE11 deficiency increases sensitivity to poly (ADP-ribose) polymerase inhibition in microsatellite unstable colorectal cancers. Cancer Res 71:2632–2642
Martin DS, Bertino JR, Koutcher JA (2000) ATP depletion + pyrimidine depletion can markedly enhance cancer therapy: fresh insight for a new approach. Cancer Res 60:6776–6783
Cepeda V, Fuertes MA, Castilha J, Alonso C, Quevedo C, Soto M, Perez JM (2006) Poly(ADP-Ribose) polymerase-1 (PARP-1). inhibitors in cancer chemotherapy. Recent Pat Anti-Cancer Drug Discov 1:39–53
Wang G, Chan CH, Gao Y, Lin HK (2012) Novel roles of Skp2 E3 ligase in cellular senescence, cancer progression, and metastasis. Chin J Cancer 31:169–177
Hsieh HY, Shieh JJ, Chen CJ, Pan MY, Yang SY, Lin SC, Chang JS, Lee AY, Chang CC (2012) Prodigiosin down-regulates SKP2 to induce p27(KIP1) stabilization and antiproliferation in human lung adenocarcinoma cells. Br J Pharmacol 166:2095–2108
Acknowledgments
This study was supported by FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo) and CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), Brazil.
Conflict of interest
The authors declare that they have no conflict of interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
da Silva, G.N., de Camargo, E.A., Sávio, A.L.V. et al. MRE11A and SKP2 genes are associated with the increased cytotoxicity induced by the synergistic effects of cisplatin and gemcitabine in bladder cancer cells. Mol Biol Rep 41, 4613–4621 (2014). https://doi.org/10.1007/s11033-014-3332-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3332-1