Abstract
AtGALK2 belongs to galactokinase of GHMP family in Arabidopsis thaliana. Two homozygous T-DNA insertion mutants (Atgalk2-1 and Atgalk2-2) of the AtGALK2 gene were identified. The AtGALK2 gene was highly expressed in flowers and roots, but less in stems, leaves and petioles. It was found that the expression of AtGALK2 gene was induced by NaCl and ABA. The two Atgalk2 mutants showed higher germination activity when treated with ABA and NaCl than wild type (Col-0). Through comparing the results of seed germination, root growth, stomatal aperture, water loss, and proline accumulation between the Atgalk2 mutants and Col-0, it was found that Atgalk2 mutants showed less sensitive to ABA than Col-0. The expression levels of ABI1, ABI2, RAB18, ABF3, RD22, RD29A, and RD29B in the Atgalk2 mutants were higher than in Col-0. However, the expression level of OST1 in the Atgalk2 mutants was lower than in Col-0. Taken together, these results suggested AtGALK2 was required for abscisic acid regulation of seed germination, root growth and gene expression, and was involved in salt and osmotic stress response in the early development stage. This study provides important clues to galactokinase activities of GHMP family in ABA signaling and plant development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The GHMP kinase family is a family of kinase enzymes. Using protein sequence analysis, the GHMP kinases were first identified in 1993 as a group of sugar kinases distinct from the hexokinase and ribokinase classes [1]. This family is named after four of the kinases including galactokinase, homoserine kinase, mevalonate kinase and phosphomevalonate kinase. These kinases make up the GHMP kinase superfamily of ATP-dependent enzymes [1–3]. The GHMP super kinase family protein involves in carbohydrates and sterols metabolism and playing an important role in eukaryote metabolism [3].
Abscisic acid (ABA) owes its names to its role in the abscission of plant leaves. In preparation for winter, it was originally believed to be involved in abscission. The plant genes for ABA biosynthesis and sequences of the pathway have been elucidated [4–6]. The ABA-mediated signalling plays an important part in plant responses to environmental stress [7, 8]. ABA is also produced in the roots in response to decreased soil water potential and other situations in which the plant may be under stress. ABA then translocates to the leaves, where it rapidly alters the osmotic potential of stomatal guard cells, causing them to shrink and stomata to close. The ABA-induced stomatal closure reduces transpiration, thus preventing further water loss from the leaves in times of low water availability [2, 9]. Seed germination is inhibited by ABA in antagonism with gibberellin. ABA also prevents loss of seed dormancy [10]. In plants, excess Gal can inhibit root or shoot growth, and cause instance deleterious physiological effects [11–13]. Therefore the galactokinase genes (GALK) may be involved to the ABA signaling.
The AtGALK2 (AT5G14470) gene is a member of the galactokinase genes in Arabidopsis. The gene chip results in Genevestigator Expression Data [14] showed that the expression of AtGALK2 was induced by ABA greatly. The 892 bp genomic DNA sequence of AtGALK2 (−888 to +3) was analyzed by online tools plantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [15], and the plant hormone and stress responsive elements were found, including Box-W1, GARE-motif (GA response element), LTR-element (low-temperature responsiveness), MYB binding site (drought Induced), and TCA-element (salicylic acid responsiveness). Therefore AtGALK2 gene may be related to hormones and environmental stresses, or play a role in hormone and stress-response pathways. To further study the biological function of AtGALK2 gene, we investigate the genes expression level in different organs, response to ABA and NaCl, and its role in ABA signaling.
Materials and methods
Plant material and growth conditions
The salk_060407 and salk_048820 T-DNA insertions mutants of AtGALK2 gene donated by ABRC (The Arabidopsis Biological Resource Center) were named Atgalk2-1 and Atgalk2-2 respectively. For in vitro culture, seeds were surface-sterilized in 75 % ethanol for 30 s, soaked by 10 % NaClO for 10 min, washed five times in sterilized water, stored at 4 °C for 3 days. Finally, these seeds were sowed on Murashige and Skoog (1962) plates containing solid medium composed of Murashige and Skoog basal salts and 1 % (w/v) Suc, solidified with 1 % (w/v) agar and the pH was adjusted to 5.7 with KOH before autoclaving. Plates were sealed and incubated in a growth chamber for ABA and NaCl treatment, seed germination and root elongation assay. The plants were routinely grown in a growth chamber under 40 % humidity, a temperature of 22 °C and with a 16-h light/8-h dark photoperiod at 100–150 μmol m−2s−1of light.
Mutant identification
To identify T-DNA insertion mutations of the AtGALK2 gene, their seedlings were screened by kanamycin firstly. Then the genomic DNAs of seedlings were isolated and used for PCR genotyping. The primers for T-DNA mutant identification were showed in Table 1. Leaves of the homozygotes were collected for RNA isolation and for further identification from gene expression level.
Abiotic stress
Plants were grown on Murashige and Skoog plates supplemented with 1.5 % Suc. After 2 weeks, Arabidopsis seedlings were treated with 100 mM NaCl or 10 μM ABA, respectively. With different time treatment, plants were collected and frozen in liquid nitrogen for RNA isolation.
Germination and root growth assays
To score seed germination, seeds were surface sterilized and plated on Murashige and Skoog solid medium with 1 % Suc, and different concentrations of ABA (0, 0.3, or 0.6 μM). To determine the sensitivity of germination to salt stress, the MS medium was supplemented with different concentrations of NaCl (0, 50, or 100 mM). Each value represents the average germination percentage of about 300 seeds at least three replicates.
Seeds were plated on 1/2 MS plates and grown for 6 day in the greenhouse. The surviving seedlings were transferred to the MS medium containing different concentrations of ABA (0, 0.1, 1, 10, or 40 μM). The root length was measured after 6 day of growth. Each value represents the average root length of about 30 seedlings at least three replicates.
Stoma aperture measurement
Stoma apertures were measured according to the procedure described by Pei [16]. The rosette leaves of the 4–5 week old Col-0 and mutants during the same growth stage were collected, and dipped into stoma opening solution (10 mM KCl, 7.5 mM iminodiacetic acid, 10 mM MES, and 10 mM Tris–HCl, pH 6.2) for 2 h to induce stoma open, then immersed for another 2 h in the same solution with different concentrations of ABA (0, 1, or 10 μ M). Stoma apertures were photographed and measured under a microscope (NICON TE2000). Data were expressed as the mean of three independent tests, and 100 stomata were measured for each test.
Assessment of the water loss rate
The water loss rate was measured as described by Shan [17]. Four fully expanded rosette leaves were collected from the 3-week-old Col-0 and mutants. After positioning them onto clean filter paper, the detached leaves were placed into a temperature controlled growth chamber at 25 °C with 60 % humidity. Fresh weight was recorded at 30 min intervals and the water loss rate was calculated using the fresh weight loss of the detached leaves. Each experiment was repeated three times.
Proline content measurement
Col-0 and mutants were treated with ABA for 3 days by spraying 0 or 100 μM ABA solution on the leaves once a day. Rosette leaves were collected and proline was extracted using the sulfosalicylic acid method [18, 19]. The proline content was measured using the absorbance at 520 nm. Each experiment was repeated for three times.
Total RNA isolation
Total RNA was isolated using an RNA easy Mini Kit (Ambiogen Biological Technology) according to the manufacturer’s instructions. First strand cDNA was synthesized using the Maxima® First cDNA Synthesis Kit (Fermentas) according to the manufacturer’s specification.
Semi-quantitative RT-PCR and quantitative RT-PCR analysis
For RT-PCR, the cDNA product was diluted for tenfold, and 1 μL of diluted cDNA was used in a 20 μL PCR reaction. PCR programs for gene expression analysis was generally performed with a 5 min denaturation at 95 °C, followed by 26 or 32 cycles consisting of the following steps: 95 °C for 30 s, 58 °C for 30 s, 72 °C for 30 s for identifying homozygous.
For quantitative RT-PCR the cDNA product was diluted 20 fold, and 2 μ L of diluted cDNA was used in a 20 μL PCR reaction. The PCR was performed using a SYBR® Green I kit (TOYOBO, Japan) in an Mx3000P PCR machine (Strata-gene, USA). The reaction started with a denaturation stage at 95 °C for 10 min, which was then followed by 40 cycles, each cycle composed of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s. Each experiment was repeated for three times. The ACTIN2 gene was used as an internal control. The relative expression level was analyzed by the Mx3000P software. The Primers for Quantitative real time PCR and Semi-quantitative RT-PCR in this study are listed in (Table 2).
Results
Identification of the T-DNA insertion mutant Atgalk2-1 and Atgalk2-2
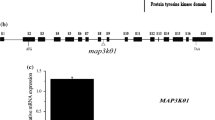
Homozygous individuals were identified by PCR and RT-PCR analyses. Sequencing of the T-DNA flanking regions in Atgalk2-1 and Atgalk2-2 showed that the insertions were located 820, or 1,627 nucleotides downstream from the ATG start codon respectively (Fig. 1a). The No. 2 line of Atgalk2-1 was identified to be homozygous (Fig. 1b). The No. 2 and No 3 line of Atgalk2-2 were identified to be homozygous (Fig. 1b). In this study, the homozygous No. 2 line plants of Atgalk2-1 and No. 3 line plants of Atgalk2-2 were selected for further research. The expressions of the AtGALK2 gene were seriously impaired in Atgalk2-1 and Atgalk2-2 seedling based on semi-quantitative RT-PCR (Fig. 1c).
Identification of the T-DNA insertion mutant of AtGALK2. a The schematic represents the AtGALK2 gene locus in the salk_060407 and salk_048820. b The screening of T-DNA insertion homozygotes of AtGALK2 gene. Lanes 1–4 were using the primers 5′ + 3′. Lanes 1′–4′ were using the primers 5′ +LBpROK2. LBpROK2 means the T-DNA left border primer. Lane M means the DNA maker. c RT-PCR analysis of AtGALK2 gene in Atgalk2-1 and Atgalk2-2 mutants, and ACTIN-2 was used as control
Expressions of the AtGALK2 gene in different organs and in response to various stresses
The expressions of ATGALK2 gene in roots, stems, stem leaves, flowers and petioles of Arabidopsis were analyzed. The results showed that AtGALK2 was expressed extremity higher in root and flower, lower in other tested organs (Fig. 2a), indicating that AtGALK2 might play a role in root and flower development. The 2-week-old Col-0 seedlings were treated with ABA (Fig. 2b) or NaCl (Fig. 2c) for different times to inspect the expression of AtGALK2 gene under different stress. The results indicated the expressions of AtGALK2 gene were induced by ABA or NaCl stress. Expression of AtGALK2 gene peaked when treated by ABA for 1 h or NaCl stress for 2 h, implying AtGALK2 gene may participate in the ABA stress signaling pathway.
Expression analysis of AtGALK2 gene in Arabidopsis. a The expression of AtGALK2 gene in different tissues of Col-0 plants. b The expression of AtGALK2 gene in 2-week-old Col-0 seedlings treated with 10 μM ABA for 0, 1, 2, or 4 h. c The expression of AtGALK2 gene in 2-week-old Col-0 seedlings treated with 100 mM NaCl for 0, 1, 2, or 4 h
AtGALK2 involved in the ABA-mediated inhibition of seed germination and root elongation
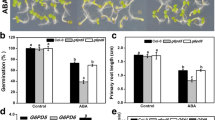
The germination ratio was tested in Murashige and Skoog (MS) solid medium containing different concentrations of ABA (0 μM, 0.3 μM, or 0.6 μM) or NaCl (0 mM, 50 mM, or 100 mM) (Fig. 3a, b, c). In the absence of exogenous ABA or NaCl, the germination rates of Atgalk2-1 and Atgalk2-2 mutants were similar to that of Col-0. With the increasing series of ABA or NaCl concentrations, Col-0 and mutant seed germination were restrained obviously. The germination rate of Col-0 was 50 %, and the germination rates of Atgalk2-1 and Atgalk2-2 were 62.5 and 64.9 % respectively treated with 0.3 μM ABA. The seed germination of Col-0 was 12.6 %, and seed germinations of Atgalk2-1 and Atgalk2-2 were 21.9 and 24.2 % treated with 0.6 μM ABA (Fig. 3a, b). The seed germination activity showed AtGALK2 T-DNA insertion mutants were less sensitive to ABA compared with the Col-0. When treated with 50 mM NaCl, the germination rates of Col-0, Atgalk2-1 and Atgalk2-2 were 77.3, 82.9, and 81.5 % respectively. When treated with 100 mM NaCl, the germination rates of Col-0, Atgalk2-1 and Atgalk2-2 were 42.3, 47.2 and 46.1 % respectively (Fig. 3a, c). The results showed that AtGALK2 T-DNA insertion mutants were more tolerant to NaCl than Col-0. To check AtGALK2 gene function in ABA signaling, 6-day-old seedlings of Col-0 and mutants were shifted to vertical MS solid plate with 0, 10, 40 μM ABA for another 6 days. The results showed that the T-DNA insertion mutants were less sensitivity to ABA than Col-0. When treated with 10 μM ABA, the T-DNA insertion mutants had more root elongation apparently than Col-0, with 40 μM ABA the root elongation almost ceased (Fig. 3d, e).
Seed germination and seedling root length of Col-0, Atgalk2-1, and Atgalk2-2 treated by exogenous ABA and NaCl. a, b, c Pictures of seed germination of Col-0, Atgalk2-1 and Atgalk2-2 grown on MS solid medium with ABA or NaCl treatment for 5 day. Approximately 300 seeds of each genotype are sowed on each plate and scored. Values are averages ± SD for three independent experiments. d, e Root length of Col-0, Atgalk2-1 and Atgalk2-2 with ABA treatment. At least 300 seedlings were measured. Values are averages ± SD for three independent experiments
The comparison of stoma aperture, water loss, and proline contents in Col-0 and Atgalk2 mutants
In order to get through drought condition, ABA is produced in plants to induce stoma closure, accumulate proline and reduce water loss [20–22]. To inspect whether AtGALK2 participate in ABA-mediated control of stoma closure, leaves with identical position were detached from 3-week-old Col-0 plants and mutants, immersed in stoma opening buffer for 2 h to induce stoma open, then treated for another 2 h with 0, 1 or 10 μM ABA. The results showed that the stoma aperture of Col-0 and mutants had a significant difference (Fig. 4a, b). The Atgalk2-1 and Atgalk2-2 showed less sensitive to ABA than Col-0 plants, and the ratio of the traverse to the longitudinal diameter of the stomata in foa1 was higher than that in Col-0 plants. These results indicated that AtGALK2 might play a positive role in ABA-mediated stoma closure. Stoma closure triggered by ABA is a crucial survival mechanism for plants to reduce water loss in response to drought stress [18, 19]. To illustrate the impact of AtGALK2 in water loss regulating, leaves of the same stage were detached and placed in a temperature controlled growth chamber, weight at 1 h intervals. In accordance with the stoma aperture results, the water-loss rate of AtGALK2 T-DNA insertion mutants were higher than that Col-0 plants (Fig. 4c), due to the larger stoma aperture, lacking the positive response of AtGALK2 protein functions to ABA. Exogenous ABA brings the plants about the accumulation of proline [23, 24]. The proline contents of Col-0 plants and T-DNA insertion mutants were tested with or not with ABA treatment. As shown in Fig. 4d, the proline was accumulated in plants treated with ABA, and there were significant differences between the Col-0 and the mutants. Proline content increased by 0.9 and 1.0 times in Atgalk2-1 and Atgalk2-2 mutant plants, and the wild-type plants increased by 0.8 times. The results suggested that Atgalk2-1 and Atgalk2-2 mutants could accumulate more proline than Col-0 plants conspicuously. Therefore Atgalk2-1 and Atgalk2-2 mutants were more tolerant to ABA and NaCl stress.
Comparison analysis of stomatal aperture, water loss, and proline accumulation between Col-0 and AtGALK2 T-DNA insertion mutants (Atgalk2-1 and Atgalk2-2). a Pictures of stomata. b Data chart of stomata aperture ratio (L/T) longitudinal diameter to the traverse diameter. c Water loss rate of seedlings measured at indicated intervals. FW is fresh weight. d Proline accumulation of the Col-0, Atgalk2-1, and Atgalk2-2
Expression analysis of ABA and stress-responsive genes
The plant hormone ABA is the major player in mediating the adaptation of the plant to stress, so the expressions of ABA and stress-responsive genes were analyzed in the Col-0 and AtGALK2 mutants in this study including the key component and repressor ABI1 and ABI2 of the ABA signaling pathway [23, 24], ABA responsive elements-binding factor ABF3 [25], activator of the ABA signaling pathway OST1 [26], and ABA and stress-responsive genes RAB18, RD22, RD29A, and RD29B [27–30]. The expressions of the above mentioned genes were induced by ABA in all the tested plants (Fig. 5). The expression levels of ABI1, ABI2, RAB18, ABF3, RD22, RD29A, and RD29B in Atgalk2-1 and Atgalk2-2 mutants were higher than that in Col-0 plants. However, the expression level of OST1 was lower than that in Col-0 plants. The results showed that the deletion of AtGALK2 altered the expressions of ABA and stress-responsive genes, and resulted in the more tolerance to ABA and NaCl, indicating that AtGALK2 possibly participates in ABA signaling pathway as a positive regulator.
Discussion
As a Galactokinase of GHMP family, AtGALK2 catalyzes the phosphorylation of α-d-galactose (Gal) in plants to avoid the excess Gal’s inhibition of root or shoot growth, and the instance deleterious physiological effects [12, 13, 31]. In another way excess Gal might affect the ABA signaling to inhibit the root or shoot growth. The AtGALK2 gene was expressed extremity higher in root and flower, lower in other tested tissues (Fig. 2a), indicating that AtGALK2 might play a role in root and flower development. The 2-week-old Col-0 seedlings were treated with ABA (Fig. 2b) or NaCl (Fig. 2c) for series time to inspect the expression of AtGALK2 gene under ABA or NaCl stress. The results indicated the expression of AtGALK2 gene were induced by ABA or NaCl stress, and expression of AtGALK2 gene peaked when treated by ABA for 1 h or NaCl stress for 2 h, implying AtGALK2 gene may participate in the ABA stress signaling pathway.
In this study, we found that the two Atgalk2 mutants showed higher germination activity when treated with ABA and NaCl than Col-0. Through comparing the results of seed germination, root growth, stomatal aperture, and water loss between the Atgalk2 mutants and Col-0 plants (Figs. 3, 4), it was found that Atgalk2 mutants showed less sensitive to ABA than the wild type. These results indicated that AtGALK2 could up-regulate the ABA signaling, because AtGALK2 could affect the ABA-regulated biological processes, such as, seed germination, root elongation, stoma closure, and water-loss. Phenotype analysis showed that ATGALK T-DNA mutants exhibited the increased tolerance to ABA and salt stress than Col-0 plants. Therefore, the ATGALK gene plays a positive role in ABA signal transduction pathway. It is well described that under stress conditions many plant species accumulate proline as an adaptive response to adverse conditions. It is generally believed that the increase in proline content following stress injury is beneficial for the plant cell [32]. In the present study, the Atgalk2-1 and Atgalk2-2 mutants can accumulate more proline than Col-0 plants treated with ABA. Therefore Atgalk2-1 and Atgalk2-2 mutants were more tolerant to ABA and NaCl stress. The more accumulated proline in ATGALK T-DNA mutants fit well with the feature of stress resistance.
The RT-PCR analysis showed that the expressions of ABI1 and ABI2 were higher in ATGALK T-DNA mutants than in Col-0 plants. The two genes regulate numerous ABA responses, such as stoma closure, osmotic water permeability of the plasma membrane, drought-induced resistance, germination and inhibition of vegetative growth and so on. The OST1 is an activator of the ABA signaling pathway and regulate numerous ABA responses. The OST1 expression was lower in ATGALK T-DNA mutants than in Col-0 plants. However, the ABF3 expression in ATGALK T-DNA mutants was higher than in Col-0 plants. These results indicated that OST and other positive ABA signaling genes were down-regulated, and therefore resulted in the accumulation of the ABF3 transcription factor. It was consistent with the phenotype of ATGALK T-DNA mutants more tolerant to ABA.
Consistent with previous results, the expressions of RD22, RD29A, RD29B and RAB18 are strongly induced by salt, drought, cold and ABA [27–30]. The expression levels of RD22, RD29A, RD29B and RAB18 were increased in ATGALK T-DNA mutants and Col-0 plants with ABA treatment. However, their expression levels in ATGALK T-DNA mutants were higher than in Col-0 plants. These results showed that ATGALK can regulate the expression level of ABA and stress-related genes, indicating that ATGALK may positively affect the ABA signaling, and plant stress response.
References
Bork P, Sander C, Valencia A (1993) Convergent evolution of similar enzymatic function on different protein folds: the hexokinase, ribokinase, and galactokinase families of sugar kinases. Protein Sci 2(1):31–40
Chaves MM, Flexas J, Pinheiro C (2009) Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot 103(4):551–560
Holden HM, Thoden JB, Timson DJ, Reece RJ (2004) Galactokinase: structure, function and role in type II galactosemia. Cell Mol Life Sci 61(19–20):2471–2484
Milborrow BV (2001) The pathway of biosynthesis of abscisic acid in vascular plants: a review of the present state of knowledge of ABA biosynthesis. J Exp Bot 52(359):1145–1164
Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Physiol 56:165–185
Seo M, Koshiba T (2002) Complex regulation of ABA biosynthesis in plants. Trends Plant Sci 7(1):41–48
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Physiol 53:247–273
Shope JC, Mott KA (2006) Membrane trafficking and osmotically induced volume changes in guard cells. J Exp Bot 57(15):4123–4131
Finkelstein R, Reeves W, Ariizumi T, Steber C (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59:387–415
Ordin L, Bonner J (1957) Effect of galactose on growth and metabolism of Avena coleoptile sections. Plant Physiol 32(3):212–215
Roberts RM, Heishman A, Wicklin C (1971) Growth inhibition and metabolite pool levels in plant tissues Fed d-glucosamine and d-galactose. Plant Physiol 48(1):36–42
Seifert GJ, Barber C, Wells B, Dolan L, Roberts K (2002) Galactose biosynthesis in Arabidopsis: genetic evidence for substrate channeling from UDP-d-galactose into cell wall polymers. Curr Biol 12(21):1840–1845
Yamamoto R, Inouhe M, Masuda Y (1988) Galactose inhibition of auxin-induced growth of mono-and dicotyledonous plants. Plant Physiol 86(4):1223–1227
Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136(1):2621–2632
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327
Pei ZM, Kuchitsu K, Ward JM, Schwarz M, Schroeder JI (1997) Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell 9(3):409–423
Shan H, Chen S, Jiang J, Chen F, Chen Y, Gu C, Li P, Song A, Zhu X, Gao H, Zhou G, Li T, Yang X (2012) Heterologous expression of the chrysanthemum R2R3-MYB transcription factor CmMYB2 enhances drought and salinity tolerance, increases hypersensitivity to ABA and delays flowering in Arabidopsis thaliana. Mol Biotechnol 51(2):160–173
Aroca R, Del MAM, Vernieri P, Ruiz-Lozano JM (2008) Plant responses to drought stress and exogenous ABA application are modulated differently by mycorrhization in tomato and an ABA-deficient mutant (sitiens). Microb Ecol 56(4):704–719
Ingram J, Bartels D (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 47:377–403
Schroeder JI, Kwak JM, Allen GJ (2001) Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410(6826):327–330
Seki M, Umezawa T, Urano K, Shinozaki K (2007) Regulatory metabolic networks in drought stress responses. Curr Opin Plant Biol 10(3):296–302
Verslues PE, Bray EA (2006) Role of abscisic acid (ABA) and Arabidopsis thaliana ABA-insensitive loci in low water potential-induced ABA and proline accumulation. J Exp Bot 57(1):201–212
Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J (1994) Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264(5164):1448–1452
Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9(5):759–771
Oh SJ, Song SI, Kim YS, Jang HJ, Kim SY, Kim M, Kim YK, Nahm BH, Kim JK (2005) Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol 138(1):341–351
Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14(12):3089–3099
Kasuga M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K (2004) A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol 45(3):346–350
Lang V, Palva ET (1992) The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 20(5):951–962
Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Transcriptional regulation of ABI3-and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol Biol 60(1):51–68
Yamaguchi-Shinozaki K, Shinozaki K (1993) The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Mol Gen Genet 238(1–2):17–25
Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG (2005) Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33(4):e36
Mattioli R, Costantino P, Trovato M (2009) Proline accumulation in plants. Plant Signal Behav 4(11):1016–1018
Acknowledgments
This research was supported by Grants from the National Natural Science Foundation of China (31071076 and 30871325), the Program for New Century Excellent Talents in University (NCET-10-0363 to X. Guo), the Excellent Youth Foundation of Hunan Province (11JJ1005), the Project Sponsored by the Scientific Research Foundation for the Returned Overseas Chinese Scholars, the State Education Ministry ([2011]1139 to X. Guo), the Ph.D. Programs Foundation of Ministry of Education of China (2014 to X. Guo), and the SIT Project of Hunan University, 2013 and 2014.
Author information
Authors and Affiliations
Corresponding author
Additional information
Qiong Zhao and Dashi Yu have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhao, Q., Yu, D., Chang, H. et al. Regulation and function of Arabidopsis AtGALK2 gene in abscisic acid response signaling. Mol Biol Rep 40, 6605–6612 (2013). https://doi.org/10.1007/s11033-013-2773-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-013-2773-2