Abstract
Oxidative stress is one of the main risk factors for asthma development. Glutathione S-transferases play an important role in antioxidant defences and may influence asthma susceptibility. In particular, GSTM1 and GSTT1 positive/null genotypes and the GSTP1*Ile105 Val polymorphism have been analyzed in a number of genetic association studies, with conflicting outcomes. Two previous meta-analyses have attempted to clarify the associations between GST genes and asthma, but these studies have also showed contrasting results. Our aim was to perform a meta-analysis that included independent genetic association studies on GSTM1, GSTP1, and GSTT1, evaluating also the effect of potential confounding variables (i.e. ethnicity, population age, and urbanization). Systematic review and meta-analysis of the effects of GST genes on asthma were conducted. The meta-analyses were performed using a fixed or, where appropriate, random effects model. The meta-analysis of the GSTM1 (n = 35), GSTT1 (n = 31) and GSTP1 (n = 28) studies suggests that no significant associations with asthma susceptibility were observed for GSTM1 and GSTP1 gene polymorphisms, whereas a significant outcome was detected for the GSTT1 positive/null genotype (pooled OR = 1.33, 95 %CI = 1.10–1.60). However, high between-study heterogeneity was identified in all the general analyses (p heterogenetity < 0.05). The stratification analysis seems to explain the heterogeneity only in few cases. This picture is probably due to the interactive process of genetics and environment that characterizes disease pathogenesis. Further studies on interactions of GST genes with the potential oxidative stress sources and with other antioxidant genes are needed to explain the role of GST enzymes in asthma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
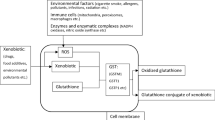
Asthma is an inflammatory disorder of the airways characterized by reversible airway obstruction and bronchial hyper-responsiveness [1]. To date, several studies have revealed that the complex interaction of cell and pro-inflammatory mediators are responsible for the pathogenesis of asthma. Among the inflammatory mediators, sources of oxidant injury are endogenous reactive oxygen species generated by cellular metabolism and by the inhalation of atmospheric pollutants [2]. These factors, which contribute to the severity and symptom exacerbation of asthma, are countered by both enzymatic and non enzymatic antioxidants, including vitamins C and E and glutathione (GSH), a major protective antioxidant in the lungs that also plays a fundamental role in the regulation of inflammatory responses [3]. Glutathione S-transferases (GSTs) catalyze the conjugation of electrophilic compounds to GSH. Their catalytic activities make these enzymes crucial to the detoxification of a wide range of endogenous and exogenous compounds [4]. Polymorphisms of genes encoding GST enzymes influence their functionality in the lungs and other organs, and this can confer genetic susceptibility to oxidative stress and, consequently, to asthma [5, 6].
For this reason, the potential role for GST polymorphisms to modulate disease and adverse responses has been the subject of numerous molecular epidemiologic studies [7–9]. The most extensively studied GST polymorphisms occurs in three isozymes: GSTM1, GSTP1, and GSTT1. Gene polymorphisms affect the activity of these enzymes: gene deletion polymorphisms occurs in the GSTM1 and GSTT1 gene, whereas a missense substitution (I105V, rs1695) characterizes GSTP1 [10].
Considering their biochemical function, the association between GST genes and asthma development risk has been thoroughly examined; published data have reported contrasting results [11, 12]. Specifically, studies have assessed the effects of GSTM1 and GSTT1 null genotypes and GSTP1 Val105 allele. Studies on GSTM1, GSTT1 and asthma have confirmed or rejected that these genes have been associated with an increased susceptibility to asthma development in children and adults. Also, for the GSTP1 gene, the Val allele reportedly protects against asthma in adults [13–15]. Conversely, in other studies, the Val105 allele seems to increase the risk of asthma in children [16, 17]. Others authors have not noted evidence of associations between GSTP1*Ile105Val polymorphism and asthma [18, 19]. All the mentioned studies have reflected upon the meta-analytical level, albeit with significant potential for improvement. Concerning the association between GST polymorphisms and asthma, only two meta-analyses have been published, with conflicting outcomes [2, 20]. Careful examination of the studies included therein reveals, however, that other eligible case–control studies have not been taken into account; additional studies have appeared, thereby increasing the amount of the available data. Furthermore, the previous meta-analyses did not analyze the effects of important confounding variables, such as ethnicity and urbanization.
The aim of our study is to perform a meta-analysis that includes genetic association studies on GST and asthma risk, analyzing the effect of ethnicity, population age, and urbanization in order to explain the heterogeneity of the results.
Materials and methods
Literature search
Eligible articles were identified through a search of the Medline and Hugenet databases through February of 2012 using combination of the following keywords: “glutathione S-transferase”, “GST”, “glutathione S-transferase M1”, “GSTM1”, “glutathione S-transferase P1”, “GSTP1”, “glutathione S-transferase T1”, “GSTT1”, “asthma”, and “asthma development”. In addition, we checked all the references of relevant reviews and eligible articles that our search retrieved. No restrictions were placed on language or type of report, and conference abstracts were also included. When multiple reports were available for a single study, only the most recent article, or the article with the largest sample size, was included. Authors of studies were contacted for further information whenever the data required by the meta-analysis were not fully reported in the article.
Meta-analysis
Case–control, cohort, and cross sectional studies with any sample size examining the association between asthma and GSTM1 null genotype, GSTT1 null genotype and GSTP1 Ile105Val polymorphism were considered eligible for this meta-analysis. For each study we collected the following data: journal name, year of publication, characteristics of the included studies (in relation to the source of cases and controls), method of ascertainment of the diagnosis (inclusion and exclusion criteria), asthma characteristics (atopic or non- atopic), demographic characteristics of the population being studied, prevalence of meaningful risk factors (smoking habits, and family history of asthma). This was done both in cases and controls. We also collected data on geographical location and ethnicity. On the basis of the information reported by the authors, we divided geographical location in urban areas or non urban areas by population estimate in accordance with demographia world urban areas: seventh annual edition [http://www.demographia.com/db-worldua.pdf].
We excluded from our analysis familiar, only-case, and occupational asthma studies. For each study frequencies of GSTP1*I105V genotypes and frequencies of GSTM1 and GSTT1 positive/null genotypes in cases and controls and crude odds ratios (ORs) were collected. A wide range of studies evaluated GSTM1 and GSTT1 as presence/absence of gene deletion, so meta-analyses of these polymorphisms were performed using a single OR (null vs. present). Results for GSTP1 Ile105Val were reported as two ORs: a dominant OR (Ile/Val + Val/Val vs. Ile/Ile) and a recessive OR (Val/Val vs. Ile/Ile + Ile/Val). Regarding GSTP1 Ile105Val polymorphisms, a sensitivity analysis was performed excluding studies where allele frequencies exhibited a significant deviation from the Hardy–Weinberg equilibrium (HWE). Statistical significance was defined as p < 0.05. The Q test and the I 2 statistic were used to investigate respectively the presence of between-study heterogeneity and the proportion of variation across studies due to heterogeneity rather than random error [21]. Possible causes of heterogeneity were investigated by subgroup analyses based on ethnicity, urbanization, and population age. For ethnicity, we considered four ethnic classes: Asian, European, Mediterranean African, and Turkish. For urbanization, the variables were defined as metropolis (>1 million inhabitants), city (500 thousand to 1 million inhabitants), and country-side (<500 thousand inhabitants). In the analyses, we used as a subgroup variable the combination of these categories, or, rather, we used a variable with four categories: 1= “Cases: Adults and Controls: Adults”, 2= “Cases: Children and Controls: Children–Adults”, 3= “Cases: Children and Controls: Adults”, 4= “Cases: Children and Controls: Children”. All statistical analyses were performed using STATA 10.1 (Stata Corp, College Station, TX, USA); the meta-analyses were performed using a fixed or, where appropriate, random effects model to estimate pooled OR. Publication bias was evaluated with the funnel plot and the Begg and Mazumdar Rank correlation test and Egger’s regression method. Then with Duvall and Tweedie’s trim and fill procedure, we tested how the effect size would shift, when the apparent bias was removed. Temporal effect was also estimated with a cumulative meta-analysis.
Results
Systemic review and meta-analysis
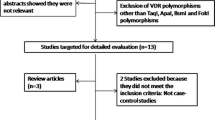
In Fig. 1, we reported the flow-chart that represents the inclusion and exclusion criteria of studies. From our initial research we identified 2,483 articles. Among those, after screening for titles and abstracts, we identified 95 studies; after cross-checking references we identified 20 additional papers so we examined and retrieved 115 papers for more detailed evaluation. 43 eligible articles were considered; we excluded occupational asthma reports, multiple reports of the same studies, family-based studies, other outcomes reports, editorials and reviews. We contacted the authors of 14 papers with no genotype data available and seven of those authors gave us the genotype data.
We included in the meta-analysis data collected from cohorts (n = 3), cross-sectional studies (n = 3), and case–control studies (n = 37). For the cohorts and cross-sectional studies we contacted authors to better define a control group (defined as group of individual without any respiratory disease), in order to equate to a case–control study and to collect data on GST gene polymorphism distribution. Association with asthma was evaluated in 35 studies on GSTM1, 31 on GSTT1, and 28 on GSTP1. In Table 1, other characteristics of the population based studies included in the meta-analyses are summarized, while genotype counts by disease outcome are reported in Supplemental Table 1 (Online Resource 1).
Meta-analyses on GSTM1 positive/null genotype
For the evaluation of the association of GSTM1 and asthma, 35 published studies were considered, a total of 6,661 affected and 17,220 non-affected individuals were included. The meta-analysis showed an increased risk of asthma associated with the GSTM1 null genotype (pooled OR = 1.12, 95 %CI = 0.99–1.26, p = 0.072; Fig. 2). This outcome, however, appears to be insignificant and large between-study heterogeneity was observed (Q = 109.17, p < 0.001; I 2 = 68.9 %). A funnel plot was performed to evaluate the publication bias of literature on GSTM1 and asthma. The shape of the funnel plot seemed asymmetrical (Supplemental Fig. 1; Online Resource 2), but the Begg and Mazumdar Rank correlation test (p = 0.551) and Egger’s regression method (bias = 1.95, t = 1.06, p = 0.299) highlighted no publication bias. Moreover, a temporal effect has been recognized in the study outcomes (Supplemental Fig. 2; Online Resource 3).
To explain the large heterogeneity in the general meta-analysis, stratification analyses were conducted. Firstly we considered ethnicity as a stratification variable. 16 studies were carried out on European populations, ten studies on Asian populations, four studies on Turkish population, and three studies evaluated the Mediterranean African population. Two studies with particular ethnic origins (a study on Iranian population [22] and a study on a mixed European/African-American population [23]) were not reported. No statistical association was observed in each ethnic group (Europe: pooled OR = 1.08, 95 %CI = 0.98–1.19; Asia: pooled OR = 1.07, 95 %CI = 0.84–1.36; Turkey: pooled OR = 1.10, 95 %CI = 0.61–1.99; Mediterranean Africa: pooled OR = 0.84, 95 %CI = 0.28-–2.52) and a high heterogeneity within each group was observed, with the exception of the European subgroup in which there was a consistency (Europe: I 2 = 24.6 %, p = 0.176; Asia: I 2 = 76.2 %, p < 0.001; Turkey: I 2 = 72.4 %, p < 0.013; Mediterranean Africa: I 2 = 88.9, p < 0.001). In the stratification analysis by urbanization, we considered 18 studies on metropolis locations, and 15 studies on country-side locations. Two studies were not reported: a study conducted in a city [24], and a study without information about the sampling location [22]. The stratification analysis by urbanization did not reveal any significant associations (metropolis: pooled OR = 1.12, 95 %CI = 0.93–1.35; country-side: pooled OR = 1.07, 95 %CI = 0.91–1.25) and highlighted the presence of a high heterogeneity within each group (metropolis: I 2 = 70.2, p < 0.001; country-side: I 2 = 66.3, p < 0.001). In the age-stratification analysis we considered 14 “Cases: Adults and Controls: Adults” studies, 11 “Cases: Children and Controls: Children” studies, four “Cases: Children and Controls: Adults” studies, and two “Cases: Children and Controls: Children–Adults” studies; we did not report four studies without age information. The OR estimated for each subgroup was not statistically significant (“Cases: Adults and Controls: Adults”: pooled OR = 1.08, 95 %CI = 0.91–1.27; “Cases: Children and Controls: Children”: pooled OR = 1.07, 95 %CI = 0.86–1.32; “Cases: Children and Controls: Adults”: pooled OR = 1.10, 95 %CI = 0.71–1.71; “Cases: Children and Controls: Children–Adults”: pooled OR = 1.45, 95 %CI = 0.89–2.37) and high heterogeneity was observed in each subgroup, with the exception of “Cases: Children and Controls: Children–Adults” (“Cases: Adults and Controls: Adults”: I 2 = 62.4 %, p = 0.001; “Cases: Children and Controls: Children”: I 2 = 66.9 %, p = 0.001; “Cases: Children and Controls: Adults”: I 2 = 73.6 %, p = 0.010; “Cases: Children and Controls: Children–Adults”: I 2 = 0.0 %, p = 0.368).
Meta-analyses on GSTT1 positive/null genotype
A total of 31 published studies on the association of GSTT1 and asthma was taken into consideration, including a total of 5,454 cases and 14,513 controls. The meta-analysis for GSTT1 shows an OR of 1.33 (95 % CI 1.10–1.60; p = 0.003) with an increased risk of asthma associated with the GSTT1 null genotype (Fig. 3). This estimate appears to be significant, but a large between-study heterogeneity was observed (Q = 118.58, p < 0.001; I 2 = 74.7 %). The funnel plot analysis revealed publication bias of literature on GSTT1 and asthma (Supplemental Fig. 3; Online Resource 4). The Begg and Mazumdar Rank correlation test (p = 0.003) and Egger’s regression method (bias = 8.76, t = 3.27 p = 0.003) consistently confirmed the visual inspection of the funnel plot. After the trim and fill procedure, there was a relevant change in overall summary estimate (OR); the procedure suggested that almost 8 studies had to be included to convert the combined p value to a non-significant value (p = 0.946) and this brought the pooled OR of 1.33–1.01. Conversely, no temporal effect seemed to be present in the study outcomes (Supplemental Fig. 4; Online Resource 5).
In the stratification analysis by ethnicity, 18 studies were carried out on European populations, four studies on Asian populations, four studies on Turkish populations, and three studies on Mediterranean African populations. The studies conducted by Saadat et al. [22] on the Iranian population and by Passos-Lima et al. [23] on European/African-American population were not reported. No significant association was observed in the ethnic groups (Europe: pooled OR = 1.18, 95 %CI = 0.98–1.43; Asia: pooled OR = 2.12, 95 %CI = 0.62–7.26; Turkey: pooled OR = 1.41, 95 %CI = 0.93–2.16; Mediterranean Africa: pooled OR = 1.73, 95 %CI = 0.60–4.96) and the studies were characterized by high heterogeneity within the subgroups, with the exception of the Turkish group (Europe: I 2 = 66.0 %, p < 0.001; Asia: I 2 = 92.5 %, p < 0.001; Turkey: I 2 = 37.2 %, p = 0.189; Mediterranean Africa: I 2 = 86.3 %, p = 0.001).
For the stratification analysis by urbanization, we considered the same studies of GSTM1. The analysis revealed significant association with high heterogeneity for “metropolis” (pooled OR = 1.65, 95 %CI = 1.12–2.42; I 2 = 85.0 %, p < 0.001) and no association with low heterogeneity for “country-side” (pooled OR = 1.05, 95 %CI = 0.91–1.21; I 2 = 27.2 %, p = 0.156).
Regarding the population age-stratification analysis, we considered 13 “Cases: Adults and Controls: Adults” studies, nine “Cases: Children and Controls: Children” studies, three “Cases: Children and Controls: Adults” studies, and two “Cases: Children and Controls: Children–Adults” studies; we did not report four studies without age information. Significant associations were observed for “Cases: Children and Controls: Children–Adults” and “Cases: Children and Controls: Adults” subgroups (“Cases: Adults and Controls: Adults”: pooled OR = 1.24, 95 %CI = 0.99–1.54; “Cases: Children and Controls: Children”: pooled OR = 1.30, 95 %CI = 0.90–1.86; “Cases: Children and Controls: Adults”: pooled OR = 0.76, 95 %CI = 0.52–1.12; “Cases: Children and Controls: Children–Adults”: pooled OR = 2.80, 95 %CI = 1.09–7.21), but only in the “Cases: Children and Controls: Adults” subgroup was no significant heterogeneity observed (“Cases: Adults and Controls: Adults”: I 2 = 64.2 %, p = 0.001; “Cases: Children and Controls: Children”: I 2 = 67.0 %, p = 0.683; “Cases: Children and Controls: Adults”: I 2 = 74.7 %, p < 0.001; “Cases: Children and Controls: Children–Adults”: I 2 = 68.6 %, p = 0.074).
Meta-analyses on GSTP1 Ile105Val polymorphism
A total of 28 published studies with 5,559 affected and 9,199 non-affected individuals was available for the meta-analysis of GSTP1*Ile105Val polymorphism. For this variant, two genetic models were considered: dominant and recessive. Regarding the general meta-analysis, no significant association and high heterogeneity were found both for the dominant (pooled OR: 0.93, 95 %CI: 0.82–1.06; Q = 59.44, p < 0.001, I 2 = 54.6 %) and for the recessive (pooled OR = 0.92, 95 %CI = 0.73–1.15, Q = 60.90, p < 0.001, I 2 = 57.3 %) genetic models (Figs. 4, 5). The funnel plots did not reveal publication bias in literature on GSTP1 and asthma in both considered genetic models (Supplemental Figs. 5 and 6; Online Resources 6, 7), and no temporal effects were observed (Supplemental Fig. 7 and 8; Online Resources 8, 9). The Begg and Mazumdar Rank correlation test (p dominant = 0.213, p recessive = 0.868) and Egger’s regression model (dominant: bias = 0.34, t = −1.90, p = 0.07; recessive: bias = 0.04, t = −0.29, p = 0.777) also confirmed this assessment.
In the stratification analysis by ethnicity, 13 studies were carried out on European populations, five studies on Asian populations, four studies on Turkish population, and four studies on the Mediterranean African populations. The single study conducted by Munoz et al. [19] on Mexican population was not reported. For the dominant model, the subgroup analysis by ethnicity highlighted no significant association and high heterogeneity for each subgroup, with the exception of the Asian and Turkish subgroups where the non-significant association were linked to low heterogeneity across studies pooled (Europe: pooled OR = 0.96, 95 %CI = 0.82–1.14, I 2 = 52.3 %, p = 0.014; Asia: pooled OR = 0.93, 95 %CI = 0.78–1.11, I 2 = 10.4 %, p = 0.349; Turkey: pooled OR = 1.11, 95 %CI = 0.77–1.59, I 2 = 55.7 %, p = 0.079; Mediterranean Africa: pooled OR = 0.59, 95 %CI = 0.27–1.29, I 2 = 82.4 %, p = 0.001). For the recessive model, a similar outcome was observed (Europe: pooled OR = 1.28, 95 %CI = 0.88–1.20, I 2 = 0.0 %, p = 0.457; Asia: pooled OR = 1.12, 95 %CI = 0.69–1.82, I 2 = 0.0 %, p = 0.578; Turkey: pooled OR = 0.89, 95 %CI = 0.32–2.44, I 2 = 83.4 %, p < 0.001; Mediterranean Africa: pooled OR = 0.52, 95 %CI = 0.17–1.61, I 2 = 83.8 %, p < 0.001). For the urbanization stratification, only one study was conducted in a city [25] and, thereby, it was not reported. 20 studies were carried out on metropolis locations and 15 studies were carried out on country-side locations. For the dominant model, the subgroup analysis by urbanization highlighted no associations and high heterogeneity within each group (metropolis: pooled OR = 0.84, 95 %CI = 0.69–1.02, I 2 = 52.7, p = 0.009; country-side: pooled OR = 1.05, 95 %CI = 0.88–1.25, I 2 = 55.2, p = 0.011). The same results were observed in urbanization stratification for the recessive genetic model (metropolis: pooled OR = 0.81, 95 %CI = 0.52–1.24, I 2 = 59.0, p = 0.003; country-side: pooled OR = 1.03, 95 %CI = 0.77–1.38, I 2 = 56.4, p = 0.003). In the stratification analysis by population age, we considered 12 “Cases: Adults and Controls: Adults” studies and 13 “Cases: Children and Cases: Children” studies. three studies were not reported because no age information was available. This stratification analysis showed no association and high heterogeneity within each group for both dominant (“Cases: Adults and Controls: Adults”: pooled OR = 0.98, 95 %CI = 0.84–1.14, I 2 = 63.3 %;p = 0.001; “Cases: Children and Controls: Children”: pooled OR = 0.92, 95 %CI = 0.76–1.13, I 2 = 41.9 %; p = 0.062) and recessive (“Cases: Adults and Controls: Adults”: pooled OR = 0.87, 95 %CI = 0.61–1.24, I 2 = 55.8 %; p = 0.009; “Cases: Children and Controls: Children”: pooled OR = 0.92, 95 %CI = 0.65–1.30, I 2 = 41.9 %; p = 0.001) models.
Discussion
GSTs are involved in many detoxification processes, and it has been hypothesized that genetic alterations of GST enzymes may affect the ability of the airways to handle toxic substances and may influence airway inflammation [12].
A number of studies has investigated the role of functional GST polymorphisms in asthma susceptibility both in children and adults, with contrasting results. To the best of our knowledge, two previous meta-analyses have attempted to clarify the association between GST genes and asthma, but these studies showed conflicting outcomes [2, 20]. Saadat and Ansari-Lari [20] stated that GSTM1 and GSTT1 null genotypes are significantly associated with asthma phenotypes, especially for non-smokers and adults. Conversely, Minelli et al. [2] reported no significant association between GSTM1, GSTP1, and GSTT1 and asthma, with high heterogeneity among the studies, even though they were stratified by asthma diagnosis.
The aim of our study was to perform a meta-analysis that included independent genetic association studies on GSTM1, GSTP1, and GSTT1, evaluating also the effect of potential confounding variables (i.e. ethnicity, population age, and urbanization).
Our results highlighted that no significant associations with asthma susceptibility were observed for GSTM1 and GSTP1 gene polymorphisms whereas significant outcomes were detected for GSTT1 positive/null genotype. However, high between-study heterogeneity was identified in all the general analyses. This heterogeneity among the genetic association studies does not ensure that these results are reliable, though [26]. Moreover, the asymmetrical distribution of the studies on GSTT1 genes in the funnel plot suggests the presence of a publication bias. In order to verify whether the heterogeneity among the studies was caused by the confounding effects of ethnicity, population age, and urbanization, we performed different stratification analyses. For GSTM1 and GSTP1 in which no associations with high heterogeneity were observed in the general analysis, the stratification revealed, in some cases, no associations with low heterogeneity, leaving most of the stratification analyses with high heterogeneity within the subgroups. Regarding GSTT1, in which a positive association was observed, a significant association with low heterogeneity was observed only in the “children versus adults” studies, whereas, in most of the other subgroups the non-significant associations were related to high heterogeneity. Therefore, the uncertainty about both the significant and non-significant results of our meta-analysis was confirmed, even after stratification analyses. Furthermore, for GSTT1 we obtained a non-significant association correcting the effect of publication bias through the trim and fill procedure. This situation is probably due to disease pathogenesis. Indeed, asthma expression is a complex process within genetics, and environment strongly impact it [27].
Our stratification analysis has tried to address confounding effects of environmental (urbanization), demographic (population age) and genetic (ethnicity) factors. Indeed, as previously reported by other authors, the inhalation of hazardous environmental factors may increase the risk for disease development or worsening of symptoms [28]. Given the multiplicity of exposures, there are many possible effects from a single pollutant, as well as from its interactions [29]. Furthermore, genetic predisposition may alter the ability of the airways to protect itself against these inhaled toxic substances from the environment [27] and genetic differences in the population structure of antioxidant enzymes may influence the genetic association between GSTs and asthma susceptibility [30]. Although our approach seems to explain a portion of this complex picture in a few cases, further studies on interactions of GST genes with environmental oxidative exposures and with other antioxidant genes are required to explain the role of GST enzymes in asthma pathogenesis.
References
Meng J, Rosenwasser LJ (2010) Unraveling the genetic basis of asthma and allergic diseases. Allergy Asthma Immunol Res 2:215–227
Minelli C, Granell R, Newson R, Rose-Zerilli MJ, Torrent M, Ring SM et al (2010) Glutathione-S-transferase genes and asthma phenotypes: a human genome epidemiology (huge) systematic review and meta-analysis including unpublished data. Int J Epidemiol 39:539–562
Nadeem A, Masood A, Siddiqui N (2008) Oxidant–antioxidant imbalance in asthma: scientific evidence, epidemiological data and possible therapeutic options. Ther Adv Respir Dis 2:215–235
Economopoulos KP, Sergentanis TN (2010) GSTM1, GSTT1, GSTP1, GSTA1 and colorectal cancer risk: a comprehensive meta-analysis. Eur J Cancer 46:1617–1631
Piacentini S, Polimanti R, Porreca F, Martínez-Labarga C, De Stefano GF, Fuciarelli M (2011) GSTT1 and GSTM1 gene polymorphisms in European and African populations. Mol Biol Rep 38:1225–1230
Polimanti R, Piacentini S, Porreca F, Fuciarelli M (2010) Glutathione S-transferase ω class (GSTO) polymorphisms in a sample from Rome (Central Italy). Ann Hum Biol 37:585–592
Piacentini S, Polimanti R, Squitti R, Ventriglia M, Cassetta E, Vernieri F et al (2012) GSTM1 null genotype as risk factor for late-onset Alzheimer’s disease in Italian patients. J Neurol Sci 317:137–140
Polimanti R, Piacentini S, Lazzarin N, Re MA, Manfellotto D, Fuciarelli M (2011) Glutathione S-transferase variants as risk factor for essential hypertension in Italian patients. Mol Cell Biochem 357:227–233
Polimanti R, Piacentini S, Lazzarin N, Vaquero E, Re MA, Manfellotto D et al (2012) Glutathione S-transferase genes and the risk of recurrent miscarriage in Italian women. Fertil Steril 98:396–400
Frova C (2006) Glutathione transferases in the genomics era: new insights and perspectives. Biomol Eng 23:149–169
Polimanti R, Piacentini S, Moscatelli B, Pellicciotti L, Manfellotto D, Fuciarelli M (2010) GSTA1, GSTO1 and GSTO2 gene polymorphisms in Italian asthma patients. Clin Exp Pharmacol Physiol 37:870–872
Piacentini S, Polimanti R, Moscatelli B, Re MA, Fuciarelli R, Manfellotto D et al (2010) Glutathione S-transferase gene polymorphisms and air pollution as interactive risk factors for asthma in a multicentre Italian field study: a preliminary study. Ann Hum Biol 37:427–439
Fryer AA, Bianco A, Hepple M, Jones PW, Strange RC, Spiteri MA (2000) Polymorphism at the glutathione S-transferase GSTP1 locus. A new marker for bronchial hyperresponsiveness and asthma. Am J Respir Crit Care Med 161:1437–1442
Abdel-Alim S, El-Masry MM, Aziz M, El-Bassiony SO, Aly AA (2007) Association of glutathione-S-transferase P1 genotypes with susceptibility to bronchial asthma in children. Arch Med Sci 3:200–207
Hanene C, Jihene L, Jamel A, Kamel H, Agnes H (2007) Association of GST genes polymorphisms with asthma in Tunisian children. Mediators Inflamm 2007:19564
Tamer L, Calikoglu M, Ates NA, Yildirim H, Ercan B, Saritas E et al (2004) Glutathione-S-transferase gene polymorphisms (GSTT1, GSTM1, GSTP1) as increased risk factors for asthma. Respirology 9:493–498
Romieu I, Ramirez-Aguilar M, Sienra-Monge JJ (2006) GSTM1 and GSTP1 and respiratory health in asthmatic children exposed to ozone. Eur Respir J 28:953–959
Safranova OG, Vavilin VA, Lyapunova AA, Makarova SI, Lyakhovich VV, Kaznacheeva LF et al (2003) Relationship between glutathione S-transferase P1 polymorphism and bronchial asthma and atopic dermatitis. Bull Exp Biol Med 136:73–75
Muñoz B, Magaña JJ, Romero-Toledo I, Juárez-Pérez E, López-Moya A, Leiva-Garcia N et al (2012) The relationship among IL-13, GSTP1, and CYP1A1 polymorphisms and environmental tobacco smoke in a population of children with asthma in Northern Mexico. Environ Toxicol Pharmacol 33:226–232
Saadat M, Ansari-Lari M (2007) Genetic polymorphism of glutathione S-transferase T1, M1 and asthma, a meta-analysis of literature. Pak J Biol Sci 10:4183–4189
Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21:1539–1548
Saadat M, Saadat I, Saboori Z, Emad A (2004) Combination of CC16, GSTM1, and GSTT1 genetic polymorphisms is associated with asthma. J Allergy Clin Immunol 113:996–998
Lima CSP, Néri IA, Lourenço GJ, Faria ICJ, Ribeiro JD, Bertuzzo CS (2010) Glutathione S-transferase mu 1 (GSTM1) and theta 1 (GSTT1) genetic polymorphisms and atopic asthma in children from Southeastern Brazil. Gen Mol Biol 33:438–441
Freidin MB, Bragina EY, Ogorodova LM, Puzyrev VP (2002) Polymorphism of the glutathione S-transferase genes (GSTT1, GSTM1) in West Siberian patients with atopic bronchial asthma. Mol Biol 36:493–496
Plutecka H, Sanak M, Diezdzina S, Mastalerz L, Szczeklik A (2006) Reactive oxygen species metabolism and allergy. Allergy Clin Immunol Int 18:158–164
Ioannidis JP (2008) Interpretation of tests of heterogeneity and bias in meta-analysis. J Eval Clin Pract 14:951–957
Kabesch M (2006) Gene by environment interactions and the development of asthma and allergy. Toxicol Lett 162:43–48
Eder W, Ege MJ, von Mutius E (2006) The asthma epidemic. New Engl J Med 355:2226–2235
Schell LM, Burnitz KK, Lathrop PW (2010) Pollution and human biology. Ann Hum Biol 37:347–366
Polimanti R, Piacentini S, Manfellotto D, Fuciarelli M (2012) Human genetic variation of cytochrome P450 superfamily: analysis of functional diversity in worldwide populations. Pharmacogenomics 13:1951–1960
Chung HW, Ahn TH, Kim SY, Kim TY, Pack D (2002) Polymorphisms of GSTM1, GSTT1, GSTP1, NATII and CYP1A1 and the susceptibility to asthma. Korean J Gen 24:259–266
Ivaschenko TE, Sideleva OG, Baranov VS (2002) Glutathione-S-transferase μ and theta gene polymorphisms as new risk factors of atopic bronchial asthma. J Mol Med 80:39–43
Vavilin VA, Makaro SI, Lyakhovich VV, Gavalov SM (2002) Polymorphic genes of xenobiotic-metabolizing enzymes associated with predisposition to bronchial asthma in hereditarily burdened and non-burdened children. Russian J Gen 38:439–445
Aynacioglu AS, Nacak M, Filiz A, Ekinci E, Roots I (2003) Protective role of glutathione S-transferase P1 (GSTP1) Val105Val genotype in patients with bronchial asthma. Br J Clin Pharmacol 57:213–217
Kabesch M, Hoefler C, Carr D, Leupold W, Weiland SK, Von Mutius E (2004) Glutathione S transferase deficiency and passive smoking increase childhood asthma. Thorax 59:569–573
Ebrahimi M, Podolskaya SV, Gorovenko NG (2004) Genetic polymorphisms of glutathione S-transferase mu1 (GSTM1) and theta1 (GSTT1) and bronchial asthma susceptibility in Ukrainian population. Iran J Biotechnol 2:230–235
Zhang YQ, Sun BY, Dai JJ, Wu SS, Zhang AP, Zhao CF et al (2004) Studies on the genetic diathesis of asthma bronchial. Yi Chuan 26:147–150
Lee Y, Hsiue T, Lee Y, Lin Y, Guo YL (2005) The association between glutathione S-transferase P1, M1 polymorphisms and asthma in Taiwanese schoolchildren. Chest 128:1156–1162
Nickel R, Haider A, Sengler C, Lau S, Niggemann B, Deichmann KA et al (2005) Association study of glutathione S-transferase P1 (GSTP1) with asthma and bronchial hyper-responsiveness in two German pediatric populations. Pediatr Allergy Immunol 16:539–541
Oh J, Kim S, Suh C, Nahm D, Park H, Lee Y et al (2005) Lack of association of glutathione S-transferase P1 Ile105Val polymorphism with aspirin-intolerant asthma. J Korean Med 20:232–236
Arbag H, Cora T, Acar H, Ozturk Sari F, Ulusoy B (2006) Lack of association between the glutathione-S-transferase genes (GSTT1 and GSTM1) and nasal polyposis. Rhinology 44:14–18
Ercan H, Birben E, Dizdar EA, Keskin O, Karaasian C, Ozge US et al (2006) Oxidative stress and genetic and epidemiologic determinants of oxidant injury in childhood asthma. J All Clin Immunol 118:97–104
Holla LI, Stejskalova A, Vasku A (2006) Polymorphisms of the GSTM1 and GSTT1 genes in patients with allergic diseases in the Czech population. Allergy 61:265–267
Imboden M, Rochat T, Brutsche MH, Schindler C, Downs S, Gerbase MW et al (2007) Glutathione-S transferase genotype increases risk of progression from bronchial hyperresponsiveness to asthma in adults. Thorax 63:322–328
Kamada F, Mashimo Y, Inoue H, Shao C, Hirota T, Doi S et al (2007) The GSTP1 gene is a susceptibility childhood asthma and the GSTM1 gene is a modifier of the GSTP1 gene. Int Arch Allergy Immunol 144:275–286
Mak JCW, Ho SP, Leung HCM, Cheung AHK, Law BKW, Sow LKY et al (2007) Relationship between glutathione S-transferase gene polymorphisms and enzyme activity in Hong Kong Chinese asthmatics. Clin Exp All 37:1150–1157
Salam MT, Lin P, Avol EL, Gauderman WJ, Gilliland FD (2007) Microsomal epoxide hydrolase, glutathione S-transferase P1, traffic and childhood asthma. Thorax 62:1050–1057
Babusikova E, Jesenak M, Kirschnerova R, Banovcin P, Dobrota D (2009) Association of oxidative stress and GST-T1 gene with childhood bronchial asthma. J Physiol Pharmacol 60:27–30
Castro-Giner F, Künzli N, Jacquemin B, Forsberg B, de Cid R, Sunyer J et al (2009) Traffic-related air pollution, oxidative stress genes, and asthma (ECHRS). Environ Health Perspect 117:1919–1924
Federova YY, Gra OA, Karunas AS, Khuzina AK, Ramazanova NN, Yuldasheva AA et al (2009) Association of polymorphisms of xenobiotic metabolism genes with childhood atopic diseases in Russian patients from Bashkortostan. Mol Biol 43:961–967
Gravina P, Angelini F, Chianca M, Valentini A, Bellincampi L, Iannini R et al (2009) GSTP1*B allele increases the risk for asthma in children. Clin Chem Lab Med 47:1451–1453
Li Y, Tseng P, Lin C, Hung C, Lin S, Su W et al (2009) NAD[P]H: quinone oxidoreductase 1, glutathione S-transferase M1, environmental tobacco smoke exposure, and childhood asthma. Mutat Res 67:53–58
Polonikov AV, Ivanov VP, Solodilova MA (2009) Genetic variation of genes for xenobiotic-metabolizing enzymes and risk of bronchial asthma: the importance of gene–gene and gene–environment interactions for disease susceptibility. J Hum Genet 54:440–449
Ozcan C, Tamer L, Ates NA, Görür K (2010) The glutathione-S-transferase gene polymorphisms (Gstt1, Gstm1, and Gstp1) in patients with non-allergic nasal polyposis. Eur Arch Otorhinolaryngol 267:227–232
Undarmaa S, Mashimo Y, Hattori S, Shimojo N, Fujita K, Miyatake A et al (2010) Replication of genetic association studies in asthma and related phenotypes. J Hum Genet 55:342–349
Abd El-Aal A, Nashar MM, El-Sissy AH (2011) Glutathione S-transferase T1 (GSTT1) and M1 (GSTM1) genes polymorphism and risk of bronchial asthma in children. Comp Clin Pathol. doi:10.1007/s00580-011-1262-z
Hersoug LG, Brash-Andersen C, Husemoen LL, Siqsqaard T, Linneberg A (2011) The relationship of glutathione-S-transferases copy number variation and indoor air pollution to symptoms and markers of respiratory disease. Clin Respir J. doi:10.1111/j.1752-699X.2011.00258.x
Mahmoud IM, Kassem HS, Abdel Wahab NH, Saad AA, Moez P (2011) The association between glutathione S-transferase P1 polymorphisms and asthma in Egyptians. Alexandria Med J 47:105–115
Tatarskyy PF, Chumachenko NG, Kucherenko AM, Gulkovskyi RV, Arabskaya LP, Smirnova OA et al (2011) Study on possible role of CYP1A1, GSTT1, GSTM1, GSTP1, NAT2 and ADRB2 genes polymorphisms in bronchial asthma development in children. Biopolym Cell 27:66–73
Karam RA, Pasha HF, El-Shal A, Rahman HMA, Gad DM (2012) Impact of glutathione-S-transferase gene polymorphisms on enzyme activity, lung function and bronchial asthma susceptibility in Egyptian children. Gene 497:314–319
Piacentini S, Polimanti R, Moscatelli B, Re MA, Manfellotto D, Fuciarelli M (2012) Lack of association between GSTM1, GSTP1 and GSTT1 gene polymorphisms and asthma in adult patients from Rome, Central Italy. J Investig Allergol Clin Immunol 22:252–256
Piacentini S, Verrotti A, Polimanti R, Giannini C, Saccucci P, Manfellotto D et al (2012) Functional polymorphisms of GSTA1 and GSTO2 genes associated with asthma in Italian children. Clin Chem Lab Med 50:311–315
Xi S, Guo L, Qi R et al (2006) Interaction of GSTM1, GSTT1 polymorphism and air pollution in asthma. J Environ Health. doi:1001-5914.0.2006-01-005
Acknowledgments
This study was supported by RSA Grant 2009 from University of Rome “Tor Vergata” and by PRIN 2009-2011 (prot. n. 200975T9EW) from MIUR.
Conflict of interest
The authors stated that there are no conflicts of interest regarding the publication of this article.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Figure 1
Funnel plot analysis to detect publication bias in study on GSTM1. Each point represents an individual study for the indicated association (TIFF 3113 kb)
Supplemental Figure 2
Cumulative forest plot to detect temporal effect in study on GSTM1 (TIFF 2324 kb)
Supplemental Figure 3
Funnel plot analysis to detect publication bias in study on GSTM1. Each point represents an individual study for the indicated association (TIFF 3113 kb)
Supplemental Figure 4
Cumulative forest plot to detect temporal effect in study on GSTT1 (TIFF 2183 kb)
Supplemental Figure 5
Funnel plot analysis to detect publication bias in study on GSTP1 (dominant genetic model). Each point represents an individual study for the indicated association (TIFF 3113 kb)
Supplemental Figure 6
Funnel plot analysis to detect publication bias in study on GSTP1 (recessive genetic model). Each point represents an individual study for the indicated association (TIFF 3113 kb)
Supplemental Figure 7
Cumulative forest plot to detect temporal effect in study on GSTP1 (dominant genetic model) (TIFF 2319 kb)
Supplemental Figure 8
Cumulative forest plot to detect temporal effect in study on GSTP1 (recessive genetic model) (TIFF 2335 kb)
Rights and permissions
About this article
Cite this article
Piacentini, S., Polimanti, R., Simonelli, I. et al. Glutathione S-transferase polymorphisms, asthma susceptibility and confounding variables: a meta-analysis. Mol Biol Rep 40, 3299–3313 (2013). https://doi.org/10.1007/s11033-012-2405-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-2405-2