Abstract
Drug metabolizing enzymes participate in the neutralizing of xenobiotics and biotransformation of drugs. Human cytochrome P450, particularly CYP1A1, CYP2C9, CYP2C19, CYP3A4 and CYP3A5, play an important role in drug metabolism. The genes encoding the CYP enzymes are polymorphic, and extensive data have shown that certain alleles confer reduced enzymatic function. The goal of this study was to determine the frequencies of important allelic variants of CYP1A1, CYP2C9, CYP2C19, CYP3A4 and CYP3A5 in the Jordanian population and compare them with the frequency in other ethnic groups. Genotyping of CYP1A1(m1 and m2), CYP2C9 (*2 and *3), CYP2C19 (*2 and *3), CYP3A4*5, CYP3A5 (*3 and *6), was carried out on Jordanian subjects. Different variants allele were determined using polymerase chain reaction-restriction fragment length polymorphism (PCR–RFLP). CYP1A1 allele frequencies in 290 subjects were 0.764 for CYP1A1*1, 0.165 for CYP1A1*2A and 0.071 for CYP1A1*2C. CYP2C9 allele frequencies in 263 subjects were 0.797 for CYP2C9*1, 0.135 for CYP2C9*2 and 0.068 for CYP2C9*3. For CYP2C19, the frequencies of the wild type (CYP2C19*1) and the nonfunctional (*2 and *3) alleles were 0.877, 0.123 and 0, respectively. Five subjects (3.16 %) were homozygous for *2/*2. Regarding CYP3A4*1B, only 12 subjects out of 173 subjects (6.9 %) were heterozygote with none were mutant for this polymorphism. With respect to CYP3A5, 229 were analyzed, frequencies of CYP3A5*1,*3 and *6 were 0.071, 0.925 and 0.0022, respectively. Comparing our data with that obtained in several Caucasian, African-American and Asian populations, Jordanians are most similar to Caucasians with regard to allelic frequencies of the tested variants of CYP1A1, CYP2C9, CYP2C19, CYP3A4 and CYP3A5.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The cytochrome P450 enzymes (CYP) play a central role in the metabolism of many therapeutic agents. Differences in the activities of these enzymes are thought to be responsible for individual variability in response and/or toxicity to numerous drugs. Among the CYP enzymes many isoforms exhibit genetic polymorphisms; examples include 1A1, 2C9, 2C19, 3A4, and 3A5. Different populations and different ethnicities differ greatly in the genotype and allele frequencies of many of the metabolizing enzymes including the CYP enzymes.
The CYP1A1 gene, located on chromosome 15q24.1. It comprises seven exons and six introns and encodes a 512–amino acid protein [1]. It plays a key role in phase I metabolism of polycyclic aromatic hydrocarbons to their ultimate DNA-binding forms [2]. A great part of the inter-individual and inter-ethnic differences in relation to xenobiotic effects is now attributable to genetic differences in their metabolism [3–5].
Three restriction fragment length polymorphic (RFLP) variants have received the most attention: m1 (MspI RFLP-CYP1A1*2A-(T6235C)-rs4646903); m2 (Bsrd1 RFLP-CYP1A1*2C-(A4889G)-rs1048943- Ile462Val); and m3 (MspI RFLP-CYP1A1*3-(T5639C)-rs4986883). The rare Val and M2 alleles of the CYP1A1 gene have been associated with increased individual cancer risk [6, 7]. Ethnic differences have been observed in the distribution of the CYP1A1 polymorphic genotypes [8] with the m1 and m2 polymorphisms being much more frequent in Asians compared to Caucasians and African-Americans [9, 10].
CYP2C9 and CYP2C19 enzymes constitute 50 %, and 16 %, respectively of the CYP2C subfamily [11]. The two genes are located on chromosome 10q24 [12]. CYP2C9 is a phase I drug-metabolizing CYP450 enzyme isoform [13], it is responsible for the catalyses of the oxidation and metabolic clearance of up to 15–20 % of clinically important drugs [14, 15] including phenytoin, warfarin, tolbutamide, losartan, and a large number of nonsteroidal anti-inflammatory-drugs [16, 17]. Changes in amino acids in this enzyme can affect its activity and produce individual variability in the concentration and/or dosage requirements of prototypic CYP2C9 substrates as warfarin and clopidogrel, respectively [18, 19].
The gene coding for the CYP2C9 enzyme is highly polymorphic, including functional variants of major pharmacogenetic importance. Thirteen variant alleles have been reported to date [13]; however, only two amino acids substitutions, the non-synonymous variants CYP2C9*2 (rs1799853-Arg144Cys) and CYP2C9*3 (rs1057910-Ile359Leu), are recognized in humans as main CYP2C9 variants [17, 20]. CYP2C9*2 and CYP2C9*3 have reduced catalytic activity compared with wild type (CYP2C9*1) [21, 22]. The frequency of CYP2C9 allelic variants has been reported to differ among Caucasian, African, and Asian populations. The allele frequencies of CY2C9*2 and CYP2C9*3 tend to be greater in Caucasian populations than in African-American and Asian populations [20–24]. The CYP2C9*2 allelic variant was not found in East Asians including Chinese and Japanese [23, 24].
CYP2C19 metabolizes several therapeutically important drugs, namely omeprazole, lansoprozole, imipramine, clopidogrel and diazepam [25, 26]. Among different variants, two alleles with separate mutations have been associated with defective enzyme in Caucasian. The first is CYP2C19*2 (rs4244285-G681A/C), which has a mutation in exon 5 causing a splice site. The other variant allele is CYP2C19*3 (rs4986893-G636A) with a point mutation in exon 4 producing a premature stop codon. The most commonly mutated allele is CYP2C19*2 in Caucasian poor metabolizers (PMs) [27] whereas, CYP2C19*3, is rare among Caucasian subjects [28].
The CYP3A locus consists of four genes, CYP3A4, CYP3A5, CYP3A7, and CYP3A43, all of which reside in a 231-kb region of chromosome 7q21.1 [29]. In adults, CYP3A4 and CYP3A5 are predominant among the four known isoforms whether in liver or intestine [30]. All genes show high degree of polymorphic expression [31]. Genetic variation within the CYP3A genes may contribute to inter-individual variability in drug metabolism. It has been suggested up to 60 % of the variability in CYP3A4 activity may be because of a genetic component [32]. The most prevalent polymorphism in CYP3A4 is (CYP3A4*1B) (rs2740574) occurs in the 5 V flanking region of the gene, it involves an A > G transition at -293 position from the transcription start site [33]. Large ethnic differences have been reported in this single nucleotide polymorphism (SNP) [30, 33].
CYP3A5 is expressed in a polymorphic manner in 10–29 % of adult livers [34, 35]. Several polymorphic variants in CYP3A5 (chromosome 7q22.1) appear to have a functional effect on CYP3A5 activity. CYP3A5*3 (rs776746) polymorphism occurs in intron 3 of CYP3A5, creating a cryptic splice site leading to the inclusion of a novel exon, and ultimately a premature stop codon, which leads eventually to very low amount of CYP3A5 protein [35, 36]. The CYP3A5 allele 6986A > G (CYP3A5*3) is associated with reduced clearance of drugs such as simvastatin, lovastatin, midazolam, cyclosporine and tacrolimus [33, 37, 38].
The expression of CYP3A5 can be reduced by quite a few other alleles in addition to CYP3A*3. The CYP3A5*6 (rs10264272) is a 14690G > A synonymous mutation (Lys–Lys) that causes the formation of a splice variant mRNA. Exon 7 is deleted, resulting in a frame shift and a truncated protein [36]. The frequency of this SNP have been found to be 17 % in African–Americans and is absent in Asians [33].
The frequencies of the important allelic variants in the CYP1A1, CYP3A4, CYP3A5 CYP2C9 and CYP2C19, genes have been studied in many ethnicities, and the data, so far, show striking interethnic variation in the distribution of these variants. However, no information is available for the Jordanian population. In the present study, we report here the allele frequencies of CYP1A1 (*2A and *2C), CYP2C9 (*2 and *3), CYP2C19 (*2 and *3), CYP3A4*1B, and CYP3A5 (*3 and *6), in a sufficiently large sample of the Jordanian population, providing a basis for future clinical studies concerning variability in the response and/or toxicity to drugs known to be substrates for these enzymes and proteins. To our knowledge this is the first genotyping study of cytochrome P450 polymorphisms in the Jordanian population reported to date.
While native Jordanians are mostly descended from people of villagers and Bedouin descent originating in the Arabian Peninsula [39], ethnically, the Jordanians represent a mixed stock. Most of the population is Arab (approximately 98 %) with 1 % of the population is Armenian, and another 1 % is Circassian. There are also small Kurd, Druze, and Chechen minorities [40, 41].
Methods
Subjects
Apparently healthy unrelated Jordanian subjects that participated in different genotyping studies were selected from many hospitals (Jordan University hospital, Al-Bashair Hospital and Royal medical service). Each subject gave a sample of about 1 ml of blood after detailed explanation of the purpose of the study; a consent form was also obtained from each subject. Genomic DNA was isolated from the blood using Wizard Genomic® (DNA purification kit) (Promega Corporation, USA). The isolated DNA samples were prepared for genotyping.
Genotyping
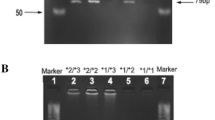
Polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) that were used to identify different single nucleotide polymorphisms (SNPs) (CYP1A1 (m1 (*2A) and m2 (*2C)), CYP2C9 (*2 and *3), CYP2C19 (*2 and *3), CYP3A4*1B, CYP3A5 (*3 and *6), primers, restriction enzymes and the condition of PCR are summarized in Table 1. Findings of the PCR-RFLP were validated by:
-
*Every time PCR reactions were done a negative control was run simultaneously. A negative control contains all PCR components except the DNA template.
-
*The sequence of the reverse, forward or both primers of (CYP2C9, CYP2C19, CYP3A4, and CYP3A5) differed from original nucleotide sequence at that region of the genomic DNA, to create an internal control site (ICS). The ICS serves to make sure that when the restriction enzyme dose not cut, it does so because of the sequence and not because it is not functioning properly. If the restriction enzyme is functional, it will always cut the PCR product at ICS irrespective of sequence of SNP.
-
*Around 15 % of all samples were repeated to confirm findings of the PCR-RFLP.
-
*The PCR-RFLP results (CYP3A4, and CYP3A5) were confirmed by direct DNA sequencing using BigDye Terminator Cycle Sequencing on 3730xl DNA sequencer (Macrogene® Co., Korea).
Statistical analysis
Allele and genotype frequencies for different alleles among Jordanian population were estimated from the results of the above PCR-RFLP test. This estimation was according to formulas reported previously [42]. Genotype and allele frequency were matched to expectation by Hardy-Weinberg Equilibrium, differences in allele frequencies between Jordanians and other ethnic populations were assessed using χ2 test [43]. A p value below 0.05 was considered statistically significant throughout the population comparisons. Table 2
The minimum sample size needed to accomplish the objectives of the study was calculated based on the following equation [44]
where N is the sample size needed; P is the reported frequency of the allele at that SNP; Q is (1 − P); Zα is the critical value at type 1 error; Zβ is the critical value at type 2 error; and Δ is the acceptable margin of error.
Based on previously reported frequencies among Caucasians and assuming α = 5 % and β = 20 % and Δ = 4 to 5 %, then the minimum sample size needed for estimation of the less-frequent allele type of CYP1A1, CYP2C9, CYP2C19, CYP3A4 and CYP3A5 was 242, 218, 143, 156, and 207 respectively.
Results
CYP1A1
The frequencies of CYP1A1 alleles among 290 Jordanian subjects were as follows; CYP1A1*1 (76.4 %), CYP1A1*2A (16.6 %) and CYP1A1*2C (7.1 %). The distribution of CYP1A1 genotypes is summarized in Table 3.
CYP2C9
The frequencies of CYP2C9 alleles among 263 Jordanian subjects were as follows; CYP2C9*1 (79.7 %), CYP2C9*2 (13.5 %), and CYP2C9*3(6.8 %). The distribution of CYP2C9 genotypes is summarized in Table 3.
CYP2C19
The frequencies of CYP2C19 alleles among 158 Jordanian subjects were as follows; CYP2C19*1 (87.65 %), CYP2C19*2 (12.35 %), and CYP2C19*3(0). The distribution of CYP2C19 genotypes is summarized in Table 3.
CYP3A4*1B
Among 173 subjects tested, 161 were homozygous for wild type allele (CYP3A4*1/*1, 93.1 %) and 12 individuals were heterozygous for variant allele (CYP3A4*1B/*1, 6.9 %). No homozygote was detected in the study samples. Thus, the allele frequency of the CYP3A4*1B variant allele among Jordanian population is 3.5 % (Table 3).
CYP3A5
The frequencies of CYP3A5 alleles among 229 Jordanian subjects were as follows; CYP3A5*1 (7.2 %), CYP3A5*3 (92.58 %), CYP3A5*6 (0.22 %). The distribution of CYP3A5 genotypes is summarized in Table 3.
All loci examined were in consistent with HWE, with the exception of one loci CYP3A5*3 (Table 3).
Discussion
CYP1A1
Ethnic differences in CYP1A1 polymorphisms have been previously reported [3–5, 45] and our results corroborate the ethnic difference in the allelic frequency of CYP1A1 between Caucasians and Africans (Table 4). The frequency of CYP1A1 mutations varies among different ethnic groups, with a high prevalence of m1 and m2 in Far East Asian populations and a low rate in Caucasians [46–48]. The m2 is found always linked with m1 [46, 48]. CYP1A1 allele *2B, containing mutations m1 and m2, was identified to place the carrier at increased risk for lung cancer [46, 49, 50] and both (*2A and *2C) are also known to increase risk for lung cancer [6, 7]. In the current study, both polymorphisms *2A/*2C were found to be carried in 34 subjects (using RFLP). All examined loci of CYP1A1 were consistent with HWE.
Our results show that the prevalence of CYP1A1*2A, and *2C were closest to the one reported for the Turkish population (Table 4). The observed genotype and allele type is quite different from Asians and Africans.
CYP2C9
The frequency of the CYP2C9*2 allele in the Jordanian population was 13.5 % which is in a range comparable with other Caucasian populations (Table 5); 8 % in American, 12.5 % in British [20], and 10.6 % in Turkish [51]. It was reported that the CYP2C9*2 allele occurs at a significantly lower frequency in the African-American population (1 %) [18]. In contrast, the CYP2C9*2 allele was reported to be absent or at least very rare in the East Asian populations [52, 53].
The CYP2C9*3 allele occurred with a frequency of 6.8 % in the Jordanian subjects. This finding is similar to that of other Caucasian populations in which the frequency of the CYP2C9*3 allele was reported to be 6 % in American, 8.5 % in British and 10.0 % in Turkish (Table 5). Lower frequencies were reported for the CYP2C9*3 variant in East Asian populations; 1.1–2.1 % in Japanese [52] and Korean [53] and the lowest frequency of CYP2C9*3 was reported in African-Americans (0.5 %) [20].
Warfarin is an oral anti-coagulant with narrow therapeutic index. It has been recommended that the initial dose and dose adjustment should be based on the genetic polymorphism of CYP2C9 in addition to VKORC1 [54]. Patients with CYP2C9*2 and *3 variants have longer times to dose stabilization and are at higher risk of serious and life-threatening bleeding [55]. The high prevalence of CYP2C9*2and *3 alleles among Jordanians clearly indicates the benefits of genetic testing of CYP2C9 in warfarin dosing, allowing physicians to identify patients at high risk for warfarin hyper responsiveness. In addition, our data could be extrapolated to other drugs metabolized by the CYP2C9.
CYP2C19
The genetic polymorphism of CYP2C19 has been shown to have the most interethnic variation with PM frequency ranges from 2 to 7 % in Caucasians, to 14–25 % in Asians [51]. In the present study, we found that the incidence of CYP2C19*2 among Jordanians (12.3 %) was similar to that found in other Caucasian populations, in Europe, and in Saudi Arabia [56]. Higher frequencies of CYP2C19*2 were reported for Asian populations [56], Africans and African Americans [51] (Table 6)
The presence of CYP2C19*3 in one Jordanian subject (0.0) was consistent with other Caucasian populations (Table 6). CYP2C19*3 has been regarded as an Asian mutation and accounted for the remaining alleles in Asian PMs [51].
The importance of genotyping CYP2C19 *2 and *3 has been highlighted by the United States Food and Drug Administration (FDA) which recommends that Clopidogrel therapy in populations with a high prevalence of those ‘‘risk’’ alleles be carefully administered [57]. Moreover, some investigators have shown associations between the CYP2C19 polymorphism and certain types of cancers (e.g., esophagus, stomach, lung and bladder cancers)[58, 59].
CYP3A4*1B
CYP3A4*1B allele frequency varies among different ethnic groups and has lower frequency among Caucasians (Table 7). Recently, CYP3A4*1B allele was found to be associated with lower CYP3A4 metabolism capacity. This lower capacity is explained by reduction of CYP3A4 expression due to presence of A-392G SNP which suggests that CYP3A4*1B diminishes the binding affinity of a transcription factor, resulting in lower transcriptional activity and therefore, lower enzyme activity [60].
The current study shows an allelic frequency of this SNP of 3.5 % among Jordanian population. Another study had been conducted among Jordanian population and found an allelic frequency of this SNP with 11.1 % [61] which is consistent with that reported previously for Caucasians (Table 4). The CYP3A4*1B allelic frequency varies among different ethnic groups: 0 % in Chinese and Taiwanese; 9.3–11 % in Hispanic Americans [33]; 2–9.6 % in Caucasians [62]; and 35–67 % in African Americans [30, 36].
There is debatable association of this variant with prostate cancer [63], chemotherapy-related leukemia [64], breast or ovarian cancer [65]. The presence of the CYP3A4*1B allele was shown to increase risk for Small Cell Lung Cancer in women [66].
CYP3A5
As mentioned previously, and in contrast to CYP3A4, the CYP3A5 enzyme is known to be expressed in only a small percentage of Caucasian individuals (10–30 %) and this has been linked to a common transition in intron 3 and results in nonfunctional protein [36].
Due to the importance of this SNP (CYP3A5*3) as a determinant for the expression of CYP3A5 gene in Caucasians, the current study estimated the allelic frequency of this SNP. In the Jordanian population the allelic frequency of this SNP was 92.58 %, which is fully consistent with that for other Caucasian populations [30, 67]. The prevalence of this SNP was 89–91 % in Caucasian-Americans; 92–94 % in European-Caucasians; 60–66 % in Hispanics, 71–75 % in East Asian populations (Chinese, Japanese, Koreans); 55–65 % in South Asians (Malay and Indians); and 29–35 % in African-Americans [33] (Table 8).
Contrary to examined HWE in all previously tested loci, we found a significant difference between the observed and the expected frequencies with regard to CYP3A5*3. The findings could be spurious. If only one sample was *1/*3 instead of *1/*1, the χ2 will change from 8.5 to 4.1 and the p value will change from 0.003 to 0.04. Alternatively, this evolution can be explained by genetic drift that occurs when the population size is limited and therefore by chance, certain alleles increase or decrease in frequency. Allele frequencies in small populations do not generally reflect those of larger populations since too small of a set of individuals cannot represent all of the alleles for the entire population [68, 69]. Only a single locus out of nine examined loci deviated from HWE.
CYP3*6 allele, which encodes for aberrantly spliced CYP3A5 enzyme was found in 0.22 % of Jordanian population resembling results found in other Caucasian populations [30, 67].
Conclusion
The current study has led to the determination of common allelic variants of a number of important metabolizing enzymes namely CYP1A1; CYP2C9; CYP2C19; CYP3A4; CYP3A5 in Jordanian population. Some of these variant alleles, to our knowledge, are being reported for the first time among the Jordanian population. The frequencies obtained are comparable to data previously reported in other populations of Caucasian origin but differ from that observed in African and Asian populations. Comparably, a mitochondrial genetics analysis revealed that Jordanians, carry genetic variants similar to other Caucasians [70]. This is of importance as it has been demonstrated in many studies that allele frequencies of the metabolic genes are not randomly distributed throughout the human population but follow diverse ethnic and/or geographic-specific patterns [4, 71–73]. The findings of current study supported by other studies all over the globe provide merits to suggest the use of genetic polymorphisms of CYP450 SNPs as markers for ethnicity and ancestral origin.
References
Weizmann Institute of Science (2011) GeneCard for protein-coding CYP1A1. http://www.genecards.org/cgi-bin/carddisp.pl?gene=CYP1A1. Accessed 5 March 2011
McManus ME, Burgess WM, Veronese ME, Huggett A, Quattrochi LC, Tukey RH (1990) Metabolism of 2-acetylaminofluorene and benzo(a)pyrene and activation of food-derived heterocyclic amine mutagens by human cytochromes P-450. Cancer Res 50:3367–3376
Munoz S, Vollrath V, Vallejos MP, Miquel JF, Covarrubias C, Raddatz A, Chianale J (1998) Genetic polymorphisms of CYP2D6, CYP1A1 and CYP2E1 in the South-Amerindian population of Chile. Pharmacogenetics 8:343–351
Garte S (1998) The role of ethnicity in cancer susceptibility gene polymorphisms: the example of CYP1A1. Carcinogenesis 19:1329–1332
Quinones L, Berthou F, Varela N, Simon B, Gil L, Lucas D (1999) Ethnic susceptibility to lung cancer: differences in CYP2E1, CYP1A1 and GSTM1 genetic polymorphisms between French Caucasian and Chilean populations. Cancer Lett 141:167–171
Shaffi SM, Shah MA, Bhat IA, Koul P, Ahmad SN, Siddiqi MA (2009) CYP1A1 polymorphisms and risk of lung cancer in the ethnic Kashmiri population. Asian Pac J Cancer Prev 10:651–656
San Jose C, Cabanillas A, Benitez J, Carrillo JA, Jimenez M, Gervasini G (2010) CYP1A1 gene polymorphisms increase lung cancer risk in a high-incidence region of Spain: a case control study. BMC Cancer 10:463
Cosma G, Crofts F, Currie D, Wirgin I, Toniolo P, Garte SJ (1993) Racial differences in restriction fragment length polymorphisms and messenger RNA inducibility of the human CYP1A1 gene. Cancer Epidemiol Biomarkers Prev 2:53–57
Surekha D, Sailaja K, Rao DN, Padma T, Raghunadharao D, Vishnupriya S (2009) Association of CYP1A1*2 polymorphisms with breast cancer risk: a case control study. Indian J Med Sci 63:13–20
Boyapati SM, Shu XO, Gao YT, Cai Q, Jin F, Zheng W (2005) Polymorphisms in CYP1A1 and breast carcinoma risk in a population-based case-control study of Chinese women. Cancer 103:2228–2235
Gerbal-Chaloin S, Pascussi JM, Pichard-Garcia L, Daujat M, Waechter F, Fabre JM, Carrere N, Maurel P (2001) Induction of CYP2C genes in human hepatocytes in primary culture. Drug Metab Dispos 29:242–251
Entrez (2011) Gene summary CYP2C19. http://www.ncbi.nlm0.nih.gov/sites/entrez?db=gene&cmd=Retrieve&dopt=full_report&list_uids=1557. Accessed June 2011
Klein TE, Chang JT, Cho MK, Easton KL, Fergerson R, Hewett M, Lin Z, Liu Y, Liu S, Oliver DE, Rubin DL, Shafa F, Stuart JM, Altman RB (2001) Integrating genotype and phenotype information: an overview of the PharmGKB project. Pharmacogenetics Research Network and Knowledge Base. Pharmacogenomics J 1:167–170
Ali ZK, Kim RJ, Ysla FM (2009) CYP2C9 polymorphisms: considerations in NSAID therapy. Curr Opin Drug Discov Devel 12:108–114
Lee CR, Goldstein JA, Pieper JA (2002) Cytochrome P450 2C9 polymorphisms: a comprehensive review of the in vitro and human data. Pharmacogenetics 12:251–263
Miners JO, Birkett DJ (1998) Cytochrome P4502C9: an enzyme of major importance in human drug metabolism. Br J Clin Pharmacol 45:525–538
Stubbins MJ, Harries LW, Smith G, Tarbit MH, Wolf CR (1996) Genetic analysis of the human cytochrome P450 CYP2C9 locus. Pharmacogenetics 6:429–439
Limdi N, Goldstein J, Blaisdell J, Beasley T, Rivers C, Acton R (2007) Influence of CYP2C9 Genotype on warfarin dose among African American and European Americans. Per Med 4:157–169
Brandt JT, Close SL, Iturria SJ, Payne CD, Farid NA, Ernest CS 2nd, Lachno DR, Salazar D, Winters KJ (2007) Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. J Thromb Haemost 5:2429–2436
Sullivan-Klose TH, Ghanayem BI, Bell DA, Zhang ZY, Kaminsky LS, Shenfield GM, Miners JO, Birkett DJ, Goldstein JA (1996) The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics 6:341–349
London SJ, Daly AK, Leathart JB, Navidi WC, Idle JR (1996) Lung cancer risk in relation to the CYP2C9*1/CYP2C9*2 genetic polymorphism among African-Americans and Caucasians in Los Angeles County, California. Pharmacogenetics 6:527–533
Yasar U, Eliasson E, Dahl ML, Johansson I, Ingelman-Sundberg M, Sjoqvist F (1999) Validation of methods for CYP2C9 genotyping: frequencies of mutant alleles in a Swedish population. Biochem Biophys Res Commun 254:628–631
Nasu K, Kubota T, Ishizaki T (1997) Genetic analysis of CYP2C9 polymorphism in a Japanese population. Pharmacogenetics 7:405–409
Wang SL, Huang J, Lai MD, Tsai JJ (1995) Detection of CYP2C9 polymorphism based on the polymerase chain reaction in Chinese. Pharmacogenetics 5:37–42
Klotz U (2006) Clinical impact of CYP2C19 polymorphism on the action of proton pump inhibitors: a review of a special problem. Int J Clin Pharmacol Ther 44:297–302
Fukasawa T, Suzuki A, Otani K (2007) Effects of genetic polymorphism of cytochrome P450 enzymes on the pharmacokinetics of benzodiazepines. J Clin Pharm Ther 32:333–341
de Morais SM, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, Goldstein JA (1994) The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J Biol Chem 269:15419–15422
De Morais SM, Wilkinson GR, Blaisdell J, Meyer UA, Nakamura K, Goldstein JA (1994) Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol Pharmacol 46:594–598
Gellner K, Eiselt R, Hustert E, Arnold H, Koch I, Haberl M, Deglmann CJ, Burk O, Buntefuss D, Escher S, Bishop C, Koebe HG, Brinkmann U, Klenk HP, Kleine K, Meyer UA, Wojnowski L (2001) Genomic organization of the human CYP3A locus: identification of a new, inducible CYP3A gene. Pharmacogenetics 11:111–121
Lamba JK, Lin YS, Schuetz EG, Thummel KE (2002) Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev 54:1271–1294
Gibson GG, Plant NJ, Swales KE, Ayrton A, El-Sankary W (2002) Receptor-dependent transcriptional activation of cytochrome P4503A genes: induction mechanisms, species differences and interindividual variation in man. Xenobiotica 32:165–206
Ozdemir V, Kalow W, Tang BK, Paterson AD, Walker SE, Endrenyi L, Kashuba AD (2000) Evaluation of the genetic component of variability in CYP3A4 activity: a repeated drug administration method. Pharmacogenetics 10:373–388
Lakhman SS, Ma Q, Morse GD (2009) Pharmacogenomics of CYP3A: considerations for HIV treatment. Pharmacogenomics 10:1323–1339
Wrighton SA, Brian WR, Sari MA, Iwasaki M, Guengerich FP, Raucy JL, Molowa DT, Vandenbranden M (1990) Studies on the expression and metabolic capabilities of human liver cytochrome P450IIIA5 (HLp3). Mol Pharmacol 38:207–213
Hustert E, Haberl M, Burk O, Wolbold R, He YQ, Klein K, Nuessler AC, Neuhaus P, Klattig J, Eiselt R, Koch I, Zibat A, Brockmoller J, Halpert JR, Zanger UM, Wojnowski L (2001) The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics 11:773–779
Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, Maurel P, Relling M, Brimer C, Yasuda K, Venkataramanan R, Strom S, Thummel K, Boguski MS, Schuetz E (2001) Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet 27:383–391
Thervet E, Anglicheau D, King B, Schlageter MH, Cassinat B, Beaune P, Legendre C, Daly AK (2003) Impact of cytochrome p450 3A5 genetic polymorphism on tacrolimus doses and concentration-to-dose ratio in renal transplant recipients. Transplantation 76:1233–1235
Zheng H, Webber S, Zeevi A, Schuetz E, Zhang J, Bowman P, Boyle G, Law Y, Miller S, Lamba J, Burckart GJ (2003) Tacrolimus dosing in pediatric heart transplant patients is related to CYP3A5 and MDR1 gene polymorphisms. Am J Transplant 3:477–483
Lowi MR (1995) Water and power: the politics of a scarce resource in the Jordan River basin. Cambridge University Press, Cambridge
Central Intelligence Agency (2012) The World Factbook. https://www.cia.gov/library/publications/the-world-factbook/fields/2075.html. Accessed 18 Feb 2012
The Royal Hashemite Court Keys to the Kingdom—The People of Jordan. http://www.kinghussein.gov.jo/people.html. Accessed 18 Feb 2012
Brooker R (ed) (2005) Genetics, analysis and principles, 2nd edn. McGraw-Hill, New York
Graph Pad Software Inc http://www.graphpad.com/quickcalcs/chisquared1.cfm. Accessed 20 February 2011
Sanford B (1990) Pharmaceutical statistics: practical and clinical application. Marcel Dekker, Inc., New York
Garte S, Gaspari L, Alexandrie AK, Ambrosone C, Autrup H, Autrup JL, Baranova H, Bathum L, Benhamou S, Boffetta P, Bouchardy C, Breskvar K, Brockmoller J, Cascorbi I, Clapper ML, Coutelle C, Daly A, Dell’Omo M, Dolzan V, Dresler CM, Fryer A, Haugen A, Hein DW, Hildesheim A, Hirvonen A, Hsieh LL, Ingelman-Sundberg M, Kalina I, Kang D, Kihara M, Kiyohara C, Kremers P, Lazarus P, Le Marchand L, Lechner MC, van Lieshout EM, London S, Manni JJ, Maugard CM, Morita S, Nazar-Stewart V, Noda K, Oda Y, Parl FF, Pastorelli R, Persson I, Peters WH, Rannug A, Rebbeck T, Risch A, Roelandt L, Romkes M, Ryberg D, Salagovic J, Schoket B, Seidegard J, Shields PG, Sim E, Sinnet D, Strange RC, Stucker I, Sugimura H, To-Figueras J, Vineis P, Yu MC, Taioli E (2001) Metabolic gene polymorphism frequencies in control populations. Cancer Epidemiol Biomarkers Prev 10:1239–1248
Cascorbi I, Brockmoller J, Roots I (1996) A C4887A polymorphism in exon 7 of human CYP1A1: population frequency, mutation linkages, and impact on lung cancer susceptibility. Cancer Res 56:4965–4969
Kawajiri K, Nakachi K, Imai K, Yoshii A, Shinoda N, Watanabe J (1990) Identification of genetically high risk individuals to lung cancer by DNA polymorphisms of the cytochrome P450IA1 gene. FEBS Lett 263:131–133
Mrozikiewicz PM, Cascorbi I, Brockmoller J, Roots I (1997) CYP1A1 mutations 4887A, 4889G, 5639C and 6235C in the Polish population and their allelic linkage, determined by peptide nucleic acid-mediated PCR clamping. Pharmacogenetics 7:303–307
Nakachi K, Imai K, Hayashi S, Watanabe J, Kawajiri K (1991) Genetic susceptibility to squamous cell carcinoma of the lung in relation to cigarette smoking dose. Cancer Res 51:5177–5180
Drakoulis N, Cascorbi I, Brockmoller J, Gross CR, Roots I (1994) Polymorphisms in the human CYP1A1 gene as susceptibility factors for lung cancer: exon-7 mutation (4889 A to G), and a T to C mutation in the 3′-flanking region. Clin Investig 72:240–248
Hamdy SI, Hiratsuka M, Narahara K, El-Enany M, Moursi N, Ahmed MS, Mizugaki M (2002) Allele and genotype frequencies of polymorphic cytochromes P450 (CYP2C9, CYP2C19, CYP2E1) and dihydropyrimidine dehydrogenase (DPYD) in the Egyptian population. Br J Clin Pharmacol 53:596–603
Kimura M, Ieiri I, Mamiya K, Urae A, Higuchi S (1998) Genetic polymorphism of cytochrome P450s, CYP2C19, and CYP2C9 in a Japanese population. Ther Drug Monit 20:243–247
Yoon YR, Shon JH, Kim MK, Lim YC, Lee HR, Park JY, Cha IJ, Shin JG (2001) Frequency of cytochrome P450 2C9 mutant alleles in a Korean population. Br J Clin Pharmacol 51:277–280
Marin-Leblanc M, Perreault S, Bahroun I, Lapointe M, Mongrain I, Provost S, Turgeon J, Talajic M, Brugada R, Phillips M, Tardif JC, Dube MP (2012) Validation of warfarin pharmacogenetic algorithms in clinical practice. Pharmacogenomics 13:21–29
Tan GM, Wu E, Lam YY, Yan BP (2010) Role of warfarin pharmacogenetic testing in clinical practice. Pharmacogenomics 11:439–448
Goldstein JA, Ishizaki T, Chiba K, de Morais SM, Bell D, Krahn PM, Evans DA (1997) Frequencies of the defective CYP2C19 alleles responsible for the mephenytoin poor metabolizer phenotype in various Oriental, Caucasian, Saudi Arabian and American black populations. Pharmacogenetics 7:59–64
Perry E (2011) Clopidogrel hyporesponsiveness and the FDA boxed warning: detection and management of patients with genetic polymorphisms. Am J Health Syst Pharm 68:529–532
Shi WX, Chen SQ (2004) Frequencies of poor metabolizers of cytochrome P450 2C19 in esophagus cancer, stomach cancer, lung cancer and bladder cancer in Chinese population. World J Gastroenterol 10:1961–1963
Bartsch H, Nair U, Risch A, Rojas M, Wikman H, Alexandrov K (2000) Genetic polymorphism of CYP genes, alone or in combination, as a risk modifier of tobacco-related cancers. Cancer Epidemiol Biomarkers Prev 9:3–28
Rodriguez-Antona C, Sayi JG, Gustafsson LL, Bertilsson L, Ingelman-Sundberg M (2005) Phenotype-genotype variability in the human CYP3A locus as assessed by the probe drug quinine and analyses of variant CYP3A4 alleles. Biochem Biophys Res Commun 338:299–305
Dawood A (2007) Polymorphisms of cytochrome P450 xenobiotic-metabolizing enzymes cyp2D6 and cyp3A4*1B and susceptability to atherosclerosis. In: Analytical toxicology science. University of Jordan, Amman, p 74
Walker AH, Jaffe JM, Gunasegaram S, Cummings SA, Huang CS, Chern HD, Olopade OI, Weber BL, Rebbeck TR (1998) Characterization of an allelic variant in the nifedipine-specific element of CYP3A4: ethnic distribution and implications for prostate cancer risk. Mutations in brief no. 191. Online. Hum Mutat 12:289
Kittles RA, Chen W, Panguluri RK, Ahaghotu C, Jackson A, Adebamowo CA, Griffin R, Williams T, Ukoli F, Adams-Campbell L, Kwagyan J, Isaacs W, Freeman V, Dunston GM (2002) CYP3A4-V and prostate cancer in African Americans: causal or confounding association because of population stratification? Hum Genet 110:553–560
Bolufer P, Collado M, Barragan E, Calasanz MJ, Colomer D, Tormo M, Gonzalez M, Brunet S, Batlle M, Cervera J, Sanz MA (2007) Profile of polymorphisms of drug-metabolising enzymes and the risk of therapy-related leukaemia. Br J Haematol 136:590–596
Spurdle AB, Goodwin B, Hodgson E, Hopper JL, Chen X, Purdie DM, McCredie MR, Giles GG, Chenevix-Trench G, Liddle C (2002) The CYP3A4*1B polymorphism has no functional significance and is not associated with risk of breast or ovarian cancer. Pharmacogenetics 12:355–366
Dally H, Edler L, Jager B, Schmezer P, Spiegelhalder B, Dienemann H, Drings P, Schulz V, Kayser K, Bartsch H, Risch A (2003) The CYP3A4*1B allele increases risk for small cell lung cancer: effect of gender and smoking dose. Pharmacogenetics 13:607–618
van Schaik RH, van der Heiden IP, van den Anker JN, Lindemans J (2002) CYP3A5 variant allele frequencies in Dutch Caucasians. Clin Chem 48:1668–1671
Hardy GH (1908) Mendelian proportions in a mixed population. Science 28:49–50
Rodriguez S, Gaunt TR, Day IN (2009) Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol 169:505–514
Gonzalez AM, Karadsheh N, Maca-Meyer N, Flores C, Cabrera VM, Larruga JM (2008) Mitochondrial DNA variation in Jordanians and their genetic relationship to other Middle East populations. Ann Hum Biol 35:212–231
London SJ, Smart J, Daly AK (2000) Lung cancer risk in relation to genetic polymorphisms of microsomal epoxide hydrolase among African-Americans and Caucasians in Los Angeles County. Lung Cancer 28:147–155
Watson MA, Stewart RK, Smith GB, Massey TE, Bell DA (1998) Human glutathione S-transferase P1 polymorphisms: relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis 19:275–280
Weber WW (1999) Populations and genetic polymorphisms. Mol Diagn 4:299–307
Aynacioglu AS, Cascorbi I, Mrozikiewicz PM, Roots I (1998) High frequency of CYP1A1 mutations in a Turkish population. Arch Toxicol 72:215–218
Song N, Tan W, Xing D, Lin D (2001) CYP 1A1 polymorphism and risk of lung cancer in relation to tobacco smoking: a case-control study in China. Carcinogenesis 22:11–16
Taioli E, Ford J, Trachman J, Li Y, Demopoulos R, Garte S (1998) Lung cancer risk and CYP1A1 genotype in African Americans. Carcinogenesis 19:813–817
Burian M, Grosch S, Tegeder I, Geisslinger G (2002) Validation of a new fluorogenic real-time PCR assay for detection of CYP2C9 allelic variants and CYP2C9 allelic distribution in a German population. Br J Clin Pharmacol 54:518–521
Scordo MG, Aklillu E, Yasar U, Dahl ML, Spina E, Ingelman-Sundberg M (2001) Genetic polymorphism of cytochrome P450 2C9 in a Caucasian and a black African population. Br J Clin Pharmacol 52:447–450
Zand N, Tajik N, Moghaddam AS, Milanian I (2007) Genetic polymorphisms of cytochrome P450 enzymes 2C9 and 2C19 in a healthy Iranian population. Clin Exp Pharmacol Physiol 34:102–105
Kudzi W, Dodoo AN, Mills JJ (2009) Characterisation of CYP2C8, CYP2C9 and CYP2C19 polymorphisms in a Ghanaian population. BMC Med Genet 10:124
Celebi A, Kocaman O, Savli H, Aygun C, Konduk BT, Senturk O, Hulagu S (2009) The prevalence of CYP2C19 mutations in Turkish patients with dyspepsia. Turk J Gastroenterol 20:161–164
Arvanitidis K, Ragia G, Iordanidou M, Kyriaki S, Xanthi A, Tavridou A, Manolopoulos VG (2007) Genetic polymorphisms of drug-metabolizing enzymes CYP2D6, CYP2C9, CYP2C19 and CYP3A5 in the Greek population. Fundam Clin Pharmacol 21:419–426
Djaffar Jureidini I, Chamseddine N, Keleshian S, Naoufal R, Zahed L, Hakime N (2011) Prevalence of CYP2C19 polymorphisms in the Lebanese population. Mol Biol Rep 38:5449–5452
Sameer AE, Amany GM, Abdela AA, Fadel SA (2009) CYP2C19 genotypes in a population of healthy volunteers and in children with hematological malignancies in Gaza Strip. Can J Clin Pharmacol 16:e156–e162
Garsa AA, McLeod HL, Marsh S (2005) CYP3A4 and CYP3A5 genotyping by Pyrosequencing. BMC Med Genet 6:19
Sayütoúlu MA, Yildiz I, Hatirnaz O, Ozbek U (2006) Common Cytochrome p4503A (CYP3A4 and CYP3A5) and Thiopurine S-Methyl Transferase (TPMT) Polymorphisms In Turkish Population. Turkish Journal of Medical Sciences. 36:11–15
Gervasini G, Vizcaino S, Gasiba C, Carrillo JA, Benitez J (2005) Differences in CYP3A5*3 genotype distribution and combinations with other polymorphisms between Spaniards and Other Caucasian populations. Ther Drug Monit 27:819–821
Veiga MI, Asimus S, Ferreira PE, Martins JP, Cavaco I, Ribeiro V, Hai TN, Petzold MG, Bjorkman A, Ashton M, Gil JP (2009) Pharmacogenomics of CYP2A6, CYP2B6, CYP2C19, CYP2D6, CYP3A4, CYP3A5 and MDR1 in Vietnam. Eur J Clin Pharmacol 65:355–363
Kudzi W, Dodoo AN, Mills JJ (2010) Genetic polymorphisms in MDR1, CYP3A4 and CYP3A5 genes in a Ghanaian population: a plausible explanation for altered metabolism of ivermectin in humans? BMC Med Genet 11:111
Quaranta S, Chevalier D, Allorge D, Lo-Guidice JM, Migot-Nabias F, Kenani A, Imbenotte M, Broly F, Lacarelle B, Lhermitte M (2006) Ethnic differences in the distribution of CYP3A5 gene polymorphisms. Xenobiotica 36:1191–1200
Hiratsuka M, Takekuma Y, Endo N, Narahara K, Hamdy SI, Kishikawa Y, Matsuura M, Agatsuma Y, Inoue T, Mizugaki M (2002) Allele and genotype frequencies of CYP2B6 and CYP3A5 in the Japanese population. Eur J Clin Pharmacol 58:417–421
Park SY, Kang YS, Jeong MS, Yoon HK, Han KO (2008) Frequencies of CYP3A5 genotypes and haplotypes in a Korean population. J Clin Pharm Ther 33:61–65
Balram C, Zhou Q, Cheung YB, Lee EJ (2003) CYP3A5*3 and *6 single nucleotide polymorphisms in three distinct Asian populations. Eur J Clin Pharmacol 59:123–126
Acknowledgments
The authors would like to thank all who participated in the study. This study was supported in part by unconditional grant from the Deanship of Scientific Research (Jordan University, Jordan). The study sponsors had no part in the study design, data collection, data analysis, data interpretations or in the writing of the manuscript. The sponsors had no role in the decision to submit the paper for publication.
Conflict of interest
None of the authors has any financial or personal relationships that could potentially be perceived as influencing our research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yousef, AM., Bulatova, N.R., Newman, W. et al. Allele and genotype frequencies of the polymorphic cytochrome P450 genes (CYP1A1, CYP3A4, CYP3A5, CYP2C9 and CYP2C19) in the Jordanian population. Mol Biol Rep 39, 9423–9433 (2012). https://doi.org/10.1007/s11033-012-1807-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-1807-5