Abstract
Stearoyl-CoA desaturase (SCD) is a multifunctional complex enzyme important in the cellular biosynthesis of fatty acids. The present study was to investigate the association of the SCD gene with milk production traits in dairy cattle. Two single nucleotide polymorphisms (SNPs) (g.6926A>G and g.8646A>G) in introns 3 and 4, and three SNPs (g.10153A>G, g.10213T>C and g.10329C>T) in exon 5 were identified with pooled DNA sequencing and genotyped using matrix-assisted laser desorption/ionization time of flight mass spectrometry assay in 752 Chinese Holstein cows. Polymorphism g.10329C>T was predicted to result in an amino acid replacement from alanine to valine in the SCD protein. With a mixed animal model, the significant associations of the five SNPs with 305-day milk, fat and protein yields and protein percentage were determined. We further demonstrated cows with heterozygous genotypes (A/G or C/T) had highest 305 day milk yield, fat yield, protein yield and lowest protein percentage. Heterozygous cows with genotype AG at the g.6926A>G locus showed the greatest milk yield (P < 0.0001), fat yield (P < 0.0001) and protein yield (P < 0.0001) among other heterozygous genotypes at any of the loci. Dominance effects of all identified SNPs on milk, fat and protein yields and protein percentage were significant. Moreover, significant allele substitution effects at g.6926A>G locus on milk yield and at g.10213T>C on protein yield were observed. Five-locus haplotypes and strong linkage disequilibrium (D′ > 0.9) between the five SNPs were also observed. The results suggest that identified polymorphisms could be potential genetic markers to improve the production performance of Chinese Holstein.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, advances of molecular genetics in the identification of chromosomal regions or loci affecting traits of economic interest have opened appealing perspectives for the improvement of milk production traits in dairy cattle. Now scientists are trying to increase the milk production without hampering the quality of milk through marker-assisted selection and breeding programs. Gene-assisted selection, the use of functional mutations directly responsible of differences in phenotypes, is at present the most powerful and efficient option of marker-assisted selection in dairy cattle breeding programs [1]. Generally mutation or polymorphism of the gene will affect gene expression [2], rate and regulation of gene transcription, or the amino acid sequence of the gene product [3]. The relationships between polymorphisms in several genes including stearoyl-CoA desaturase (SCD) and dairy traits have already been tested in the several independent studies [4–7]. As for the bovine SCD gene, the published studies on single nucleotide polymorphisms (SNPs) are from Mele et al. [8] and Moioli et al. [9] reported the existence of SNPs in the SCD gene with significant influence on the milk fat composition in dairy cattle. It has been hypothesized that diet-independent variations in CLA content of milk fat may be due to differences in mammary SCD activity, associated with either regulation of expression and polymorphism of SCD, or differences in downstream factors that would affect interactions between enzymes and substrates (e.g., phosphorylation) [10]. SCD is a key rate-limiting enzyme in the biosynthesis of monounsaturated fatty acids (FAs) by insertion of a cis-double bond in the D9 position (between carbons 9 and 10 atoms) of methylene-interrupted fatty acyl-CoA substrates. This oxidative reaction is catalyzed by the SCD and involves cytochrome b5, NADP (H)-dependent cytochrome b5 reductase and molecular oxygen [11–13].
Because of biological significance on the quantitative traits SCD is considered as an excellent candidate gene for QTL analysis. QTL located on BTA26 affecting milk, fat and protein yields and milk composition in dairy cattle have been reported in some independent studies [14–18]. Several genes involved in lipid metabolism that are located on BTA26 including SCD, LPF (gastric lipase), or GPAM (glycerol-3-phosphate acyltransferase mitochondrial) have been suggested as possible candidate genes harboring the functional mutation detected by the QTL analysis [19]. The authors also concluded, the microsatellites used for the study were close to these genes seem to exclude the possibility that the position of the SCD locus matches the location of any of the QTL affecting milk production traits found on this chromosome.
The SCD gene has been cloned and characterized in a number of mammalian species including rodents, ovine, porcine, bovine and human [20–23]. Bovine SCD has been localized to chromosome 26q21 [20]. Northern blot analysis provides evidence for the existence of a single SCD transcript of approximately 5 kb with an unusually long 3′UTR in bovine mammary gland and adipose tissues [20, 24]. The bovine SCD is approximately 17 kb, comprises six exons and five introns (GenBank accession no. AY241932 and AY241933) and contains an open reading frame of 1,080 nucleotides coding for a protein of 359 amino acids and the 3′UTR has 3,884 nucleotides [25]. According to the biosynthesis function and QTL effects on milk fat, it is reasonable to hypothesize that SCD gene may also affect the production traits such as milk yield, fat yield, and protein yields that represent the main breeding goals in current selection schemes of dairy cattle. In the present study, the SCD gene was chosen as a positional candidate gene for milk production traits, based upon its biological function and genomic location. We screened its full-length coding region for SNPs and evaluated the genetic effects of polymorphisms on milk production traits in Chinese Holstein.
Materials and methods
Experimental animal and phenotypic data
A total of 752 Chinese Holstein cows were chosen from Beijing Dairy Cattle Center and Beijing Sanyuan Lvhe Dairy Farming Center in the Beijing region, China, including 14 sire families with 30–153 daughters from each sire. Performance data for five milk production traits (i.e. milk yield, fat yield, protein yield, fat percentage and protein percentage over 305 days) were collected from the Dairy Data Processing Center, Dairy Association of China. Genomic DNA was extracted from whole blood samples of the cows using a TIANamp Genomic DNA kit (TianGen, Beijing, China) according to the manufacturer’s instructions and frozen semen of the 14 sires by a standard phenol–chloroform method.
Detection and genotyping of the polymorphisms
A DNA pool (50 ng/μl/bull) was constructed from the 14 sires. Six pair of primers (Table 1) were designed to amplify all exons (exons 1–6) and their partial flanking intronic sequences based on the reference sequence of the bovine SCD gene (GenBank accession no. AY241932) with Primer3 web Program (v.0.4.0) in order to identify SNPs. PCR amplifications for pooled DNA from 14 sires were performed in a final reaction volume of 25 μl consisting of 50 ng genomic DNA, 0.5 μl of each primer, 2.5 μl 10× PCR buffer, 2.5 mM each of dNTP, and 1 U of Taq DNA polymerase (TaKaRa, Dalian, China). The PCR protocol was 5 min at 94 °C for initial denaturing followed by 34 cycles at 94 °C for 30 s; 56 °C for 30 s; 72 °C for 30 s; a final extension at 72 °C for 7 min for all the primers. Then, 40 μl of each PCR product from the pooled DNA was sequenced using the ABI3730XL (Applied Biosystems, Foster City, CA). Both forward and reverse primer sequences were then aligned using the ClustalW program to determine the presence of genetic polymorphism. Matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS, Squenom MassARRAY®, Bioyong Technologeies Inc. HK) assay was used for individual genotyping of 752 Chinese Holstein cows.
Statistical analysis
With tracing back of pedigree information for three generations, the total number of animals included in the analysis was 2,212. For the association studies, the traits of interest were analyzed using several statistical programs e.g. MATLAB (http://www.mathworks.com) version 7.11.0.584 (R2010b) for the kinship matrix (A-matrix), POPGENE (http://www.ualberta.ca/~fyeh/) version 1.32 for Hardy–Weinberg equilibrium, Haploview (http://www.broadinstitute.org/haploview/haploview) version 4.2 for linkage disequilibrium and haplotypes analysis. The mixed procedure of SAS 9.1.0 software (SAS OnlineDoc® 9.1.0. Cary, NC: SAS Institute Inc. USA) with the following animal model [26] was used to estimate the effects of SCD polymorphic genotypes on the milk production traits. Each trait was analysed separately and each polymorphism was also fitted separately in the model
where Y is the phenotypic data of each trait of cows, μ is the general mean, hys is the herd-year-season effect, L is the fixed effect of lactation, G is the fixed effect of polymorphism genotypes, α i is the residual polygenic effect, e is the random residual. The difference between the effects of the polymorphism genotypes were compared using multiple t test with Bonferroni correction in which the significance level of the multiple tests was equal to the significant level of single test divided by number of tests. Here there are three levels of genotype for each trait, thus three t tests needed to be performed, so the Bonferroni corrected significance levels of 0.05/3 = 0.0167 and 0.01/3 = 0.0033 were used.
The equation of Falconer and Mackay [27] was fitted to estimate the additive (a), dominance (d) and allele substitution (α) effects, i.e. a = (AA − BB)/2, d = AB − (AA + BB)/2 and α = a + d(q − p), where AA or BB indicates the two homozygous genotypes, AB indicates heterozygous genotype, p and q are the allele frequencies of corresponding loci.
Results and discussion
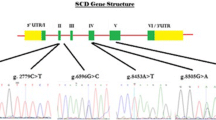
Through sequence analysis of the pooled DNA samples, five SNPs in SCD gene were discovered in this study, of which, two (g.6926A>G and g.8646A>G) were found in introns 3 and 4 respectively and another three (g.10153A>G, g.10213T>C and g.10329C>T) in exon 5 (Fig. 1). The genotypic and allelic frequencies and Hardy–Weinberg equilibrium test (χ2) are summarized in Table 2. χ2 Test showed that all genotypic frequencies in the population were in Hardy–Weinberg equilibrium (P > 0.05) (Table 2), indicating that selection pressure (mainly on production traits) did not have a large influence on the genotypic frequencies of these sites. Five-locus haplotypes are observed as shown in Table 3. The two haplotypes [AAATC] and [GGGCT] were detected in this study and the frequencies were 66 and 34 % respectively. The linkage disequilibrium between the five SNPs was estimated, and the results (Fig. 2) showed the five SNPs were in linkage disequilibrium (D′ > 0.9).
In Table 4, association analysis demonstrated that cows with genotype AG at the g.6926A>G locus showed the greatest 305-day milk yield (P < 0.0001), fat yield (P < 0.0001) and protein yield (P < 0.0001) among genotypes with 354, 12 and 7 kg more milk, fat and protein yield respectively compared with GG cows. This SNP had no effect on fat percentage. Moreover, the protein percentage of AA or GG cows was significantly greater than that of AG cows, with a difference of about 0.029 and 0.045 kg, respectively. Cows with heterozygous genotype AG at locus g.8646A>G and g.10153A>G had highest milk (P < 0.0001), fat (P < 0.05) and protein (P < 0.05) yields than the ones with homozygous AA and GG, whereas genotype GG (at both locus) showed highest protein percentage (P < 0.0001). The SNPs g.10213T>and g.10329C>T have significant effects on milk, fat and protein yields as well. Cows with CT genotypes at g.10213T>C and g.10329C>T locus had highest milk yield (P < 0.001), fat yield (P < 0.05) and protein yield (P < 0.0001; P < 0.05) whereas lowest protein percentage (P < 0.0001; P < 0.05) than in those with TT or CC respectively. However, no significant association with fat percentage was observed. We further demonstrated that dominance effects of all identified SNPs on milk yield (P < 0.001), fat yield (P < 0.001), protein yield (P < 0.001), and protein percentage (P < 0.001) were significant (Table 5). Additive effects were not significant for all of the traits whereas the significant allele substitution effects at g.6926A>G locus on milk yield (P < 0.05) and at g.10213T>C locus on protein yield (P < 0.05) were observed. As a whole, for all five loci, heterozygous genotypes led to much better performance than the homozygous genotypes for milk yield fat yield and protein yield, while the homozygous genotypes had slightly higher protein percentage and fat percentage, and differences between the two homozygous genotypes were little.
SCD is a multifunctional complex enzyme in the cellular biosynthesis of FAs. The SCD has been suggested as a candidate gene affecting milk FA profile [8, 9, 19], the FA profile of carcass fat [25, 28] and a fundamental aspect of milk nutritional quality [29, 30]. In our study, to evaluate the effect of SNPs, 752 cows were genotyped by MALDI-TOF array. The average genotyping success rate was 91.6 %. In contrast to Taniguchi et al. [25], in the current study a strong linkage disequilibrium (D′ > 0.9) was found among the SNPs; only polymorphism g.10329C>T was predicted to result in an amino acid replacement from alanine into valine in the SCD protein. The C allele was identified as the ancestral allele. As a result, similar significant associations on milk production traits were observed in several cases. This amino acid replacement considered in this study was in agreement with previous reports on the different cattle populations [8, 9, 19, 25, 28, 31, 32]. Strong LD between the analyzed three SNPs of exon 5 (D′ > 0.9) were also detected by Milanesi et al. [33], with correlation values (r 2) between g.10213T>C and g.10329C>T and between g.10153A>G and g.10213T>C (they named the SNPs A702G, T762C and T878C) reported as 0.98 and 0.65, respectively, indicating a higher recombination frequency between the latter pair of SNP. Our results also were in agreement with these results. The study carried out by Milanesi et al. [33] on Italian cattle breed reported that in the 11 breeds three-locus five haplotypes had been reconstructed. The most frequent haplotypes, ATC (55.6 %) and GCT (34.8 %), were present in all the breeds studied. These two haplotypes in the exon 5 also reported in the Japanese Black [25] and in the Italian Holstein breeds [34]. However, literature data on six-locus haplotypes frequency were not available for comparison with our results.
Higher frequency of the C allele than the T allele at position g.10329C>T was reported for Italian Holsteins (0.57) [8], Valdostana (0.65), Jerseys (0.94) [9], Grey Alpine (0.89) [34] and Japanese Black cattle (0.59) [25]. The allele frequencies of the present study at this locus were consistent with these studies. The higher C allele frequency in the Chinese Holstein population may have resulted from long term selective breeding for improving the milk production or other associated traits, or from drift. In contrast to Milianesi et al. [33], in the current study A (0.66) and T (0.66) allele were found to be more frequent at position g.10153A>G and g.10213T>C respectively.
A study was carried out by Macciotta et al. [35], who claimed that cows with homozygous genotype CC at the SCD locus (g.10329C>T) producing more milk (about 2 kg/day) and protein (about 0.07 kg/day) compared with TT cows; heterozygous cows were in an intermediate position but our results disagree with their reported literature. The results of the present experiment were consistent with previous results of Conte et al. [34] who had shown that this SNP had been associated with higher SCD (g.10329C>T) activity and fat concentration in milk in Italian Holstein.
In contrast to the results of Taniguchi et al. [25] on FA composition of carcass fat in cattle, Mele et al. [8] found a significant association between SCD genotypes and cis-9 C18:1, cis-9 C14:1, and total MUFA contents of milk. In particular, the homozygous TT genotype was associated with 9.3, 37.9, and 11.7 % more MUFA, cis-9 C14:1 and cis-9 C18:1, respectively, when compared with homozygous CC animal whereas Moioli et al. [9] did not find any association in CLA, polyunsaturated FA, and MUFA in either Valdostana or Jerseys breed. Taniguchi et al. [25] also reported that heterozygous Val/Ala (C/T) animals had higher C16:1/C16:0 in intramuscular fat than did homozygous Val (TT) animals. Another study by Conte et al. [33] on Italian brown cattle reported a significant association between SCD1 genotypes and milk FA C14:1 cis-9 and DI 14 (P ≤ 0.01). In particular, the CC genotype was associated with 18.3 and 20.6 % more C14:1 cis-9 and DI 14, respectively, compared with the TT genotype.
This seemingly paradoxical result could be explained by several reasons, e.g. the genetic background, linkage disequilibrium, artificial selection for other traits and paternal effect. Generally the effect of polymorphism will differ across different populations or breed because of specific genetic backgrounds. Chinese Holsteins cattle originated from cross-breeding between native cow (Chinese Yellow cattle) and American or European Holsteins (pure-bred bull) over the past 100 years. Continuous import of Holstein sires, semen and embryos which were directly used in AI or via crosses with Chinese Holstein cows through planned mating to increase the milk production and other traits [36]. Another possible reason for the conflicting findings between different studies could be that some of the studies were based on relatively small samples and thus the results might be involved in relatively large sampling errors.
In addition, two SNPs in introns 3 and 4 of SCD gene were detected in this study. Although, intron was not the sequence for coding protein but it played an important regulating effect in gene expression, regulation [2] and transcription and mRNA splicing [3]. Further study is needed to explore whether the detected mutations affect gene expression or not. In conclusion, our findings showed that SCD gene polymorphisms are associated with milk production traits in Chinese Holstein population; however, heterozygous cows yielded more milk, fat and protein. The results suggest that SCD is a candidate gene that influences milk production traits, and provides potentially useful information for in dairy breeding programs. The identified polymorphisms could be potential genetic markers to improve the production performance of Chinese Holstein. Further studies using long term production data and in vitro biological analysis should be conducted in order to check the effects of such polymorphisms and validate its function on milk production traits.
References
Dekkers JCM (2004) Commercial application of marker-and gene-assisted selection in livestock: strategies and lessons. J Anim Sci 82(13 Electronic Supplement 1):E313
Nott A, Meislin SH, Moore MJ (2003) A quantitative analysis of intron effects on mammalian gene expression. RNA 9(5):607–617
Zan L, Zhang J, Liu X (2007) Association study on AGPAT6 intron3 polymorphism and milk performance of dairy cattle. Sci Agric Sin 40(7):1498–1503
Grisart B, Coppieters W, Farnir F, Karim L, Ford C, Berzi P, Cambisano N, Mni M, Reid S, Simon P (2002) Positional candidate cloning of a QTL in dairy cattle: identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res 12(2):222–231
Hayes B, Goddard ME (2001) The distribution of the effects of genes affecting quantitative traits in livestock. Genet Sel Evol 33(3):209–230
Khatib H, Monson RL, Schutzkus V, Kohl DM, Rosa GJM, Rutledge JJ (2008) Mutations in the STAT5A gene are associated with embryonic survival and milk composition in cattle. J Dairy Sci 91(2):784–793
Khatib H, Zaitoun I, Wiebelhaus-Finger J, Chang YM, Rosa GJM (2007) The association of bovine PPARGC1A and OPN genes with milk composition in two independent Holstein cattle populations. J Dairy Sci 90(6):2966–2970
Mele M, Conte G, Castiglioni B, Chessa S, Macciotta NP, Serra A, Buccioni A, Pagnacco G, Secchiari P (2007) Stearoyl-coenzyme A desaturase gene polymorphism and milk fatty acid composition in Italian Holsteins. J Dairy Res 90(9):4458–4465. doi:10.3168/jds.2006-617
Moioli B, Contarini G, Avalli A, Catillo G, Orru L, De Matteis G, Masoero G, Napolitano F (2007) Short communication: effect of stearoyl-coenzyme A desaturase polymorphism on fatty acid composition of milk. J Dairy Res 90(7):3553–3558. doi:10.3168/jds.2006-855
Peterson DG, Kelsey JA, Bauman DE (2002) Analysis of variation in cis-9, trans-11 conjugated linoleic acid (CLA) in milk fat of dairy cows. J Dairy Res 85(9):2164–2172. doi:10.3168/jds.S0022-0302(02)74295-1
Enoch HG, Catala A, Strittmatter P (1976) Mechanism of rat liver microsomal stearyl-CoA desaturase. Studies of the substrate specificity, enzyme-substrate interactions, and the function of lipid. J Biol Chem 251(16):5095
Shimakata T, Mihara K (1972) Reconstitution of hepatic microsomal stearoyl-coenzyme A desaturase system from solubilized components. J Biochem 72(5):1163–1174
Strittmatter P, Spatz L, Corcoran D, Rogers MJ, Setlow B, Redline R (1974) Purification and properties of rat liver microsomal stearoyl coenzyme A desaturase. Proc Natl Acad Sci USA 71(11):4565
Boichard D, Grohs C, Bourgeois F, Cerqueira F, Faugeras R, Neau A, Rupp R, Amigues Y, Boscher MY, Levéziel H (2003) Detection of genes influencing economic traits in three French dairy cattle breeds. Genet Sel Evol 35(1):77–102
Druet T, Fritz S, Boichard D, Colleau JJ (2006) Estimation of genetic parameters for quantitative trait loci for dairy traits in the French Holstein population. J Dairy Res 89(10):4070–4076. doi:10.3168/jds.S0022-0302(06)72451-1
Jiang Z, De S, Garcia MD, Griffin KB, Wu XL, Xiao Q, Michal JJ, Sharma BS, Jansen GB (2005) An independent confirmation of a quantitative trait locus for milk yield and composition traits on bovine chromosome 26. J Anim Breed Genet 122(4):281–284
Plante Y, Gibson JP, Nadesalingam J, Mehrabani-Yeganeh H, Lefebvre S, Vandervoort G, Jansen GB (2001) Detection of quantitative trait loci affecting milk production traits on 10 chromosomes in Holstein cattle. J Dairy Res 84(6):1516–1524. doi:10.3168/jds.S0022-0302(01)70185-3
Viitala SM, Schulman NF, de Koning DJ, Elo K, Kinos R, Virta A, Virta J, Maki-Tanila A, Vilkki JH (2003) Quantitative trait loci affecting milk production traits in Finnish Ayrshire dairy cattle. J Dairy Res 86(5):1828–1836. doi:10.3168/jds.S0022-0302(03)73769-2
Gautier M, Barcelona RR, Fritz S, Grohs C, Druet T, Boichard D, Eggen A, Meuwissen TH (2006) Fine mapping and physical characterization of two linked quantitative trait loci affecting milk fat yield in dairy cattle on BTA26. Genetics 172(1):425–436. doi:10.1534/genetics.105.046169
Bernard L, Leroux C, Hayes H, Gautier M, Chilliard Y, Martin P (2001) Characterization of the caprine stearoyl-CoA desaturase gene and its mRNA showing an unusually long 3′-UTR sequence arising from a single exon* 1. Gene 281(1–2):53–61
Miyazaki M, Jacobson MJ, Man WC, Cohen P, Asilmaz E, Friedman JM, Ntambi JM (2003) Identification and characterization of murine SCD4, a novel heart-specific stearoyl-CoA desaturase isoform regulated by leptin and dietary factors. J Biol Chem 278(36):33904
Ward RJ, Travers MT, Richards SE, Vernon RG, Salter AM, Buttery PJ, Barber MC (1998) Stearoyl-CoA desaturase mRNA is transcribed from a single gene in the ovine genome1. Biochim Biophys Acta Lipids Lipid Metab 1391(2):145–156
Zhang L, Ge L, Parimoo S, Stenn K, Prouty SM (1999) Human stearoyl-CoA desaturase: alternative transcripts generated from a single gene by usage of tandem polyadenylation sites. Biochem J 340(Pt 1):255–264
Chung M, Ha S, Jeong S, Bok J, Cho K, Baik M, Choi Y (2000) Cloning and characterization of bovine stearoyl CoA desaturase1 cDNA from adipose tissues. Biosci Biotechnol Biochem 64(7):1526–1530
Taniguchi M, Utsugi T, Oyama K, Mannen H, Kobayashi M, Tanabe Y, Ogino A, Tsuji S (2004) Genotype of stearoyl-coA desaturase is associated with fatty acid composition in Japanese Black cattle. Mamm Genome 15(2):142–148
Lynch M, Walsh B (1997) Genetics and analysis of quantitative traits. Sinauer Associates, Inc., Sunderland
Falconer DS, Mackay TFC (1996) Introduction to quantitative genetics, 4th edn. Longman Scientific and Technical, New York
Zhang S (2005) Association of genetic variation to healthfulness of beef. Lowa State University Animal Industry Report 2005. www.ans.iastate.edu/report/air/2005.pdf. Accessed 12 Dec 2009
Bauman DE, Mather IH, Wall RJ, Lock AL (2006) Major advances associated with the biosynthesis of milk. J Dairy Sci 89(4):1235–1243
Soyeurt H, Dardenne P, Gillon A, Croquet C, Vanderick S, Mayeres P, Bertozzi C, Gengler N (2006) Variation in fatty acid contents of milk and milk fat within and across breeds. J Dairy Sci 89(12):4858–4865
Kgwatalala PM, Ibeagha-Awemu EM, Hayes JF, Zhao X (2007) Single nucleotide polymorphisms in the open reading frame of the stearoyl-CoA desaturase gene and resulting genetic variants in Canadian Holstein and Jersey cows. DNA Seq 18(5):357–362. doi:10.1080/10425170701291921
Schennink A, Heck JM, Bovenhuis H, Visker MH, van Valenberg HJ, van Arendonk JA (2008) Milk fatty acid unsaturation: genetic parameters and effects of stearoyl-CoA desaturase (SCD1) and acyl CoA: diacylglycerol acyltransferase 1 (DGAT1). J Dairy Res 91(5):2135–2143. doi:10.3168/jds.2007-0825
Milanesi E, Nicoloso L, Crepaldi P (2008) Stearoyl CoA desaturase (SCD) gene polymorphisms in Italian cattle breeds. J Anim Breed Genet 125(1):63–67. doi:10.1111/j.1439-0388.2007.00697.x
Conte G, Mele M, Castiglioni B, Serra A, Viva M, Chessa S, Pagnacco G, Secchiari P (2006) Relationship between bovine SCD polymorphism locus and mammary gland desaturation activity. In: 8th world congress on genetics applied to livestock production, Belo Horizonte, Minas Gerais, Brazil, 2006. Instituto Prociancia, pp 22–37. URL: http://www.wcgalp8.org.br
Macciotta NP, Mele M, Conte G, Serra A, Cassandro M, Dal Zotto R, Borlino AC, Pagnacco G, Secchiari P (2008) Association between a polymorphism at the stearoyl CoA desaturase locus and milk production traits in Italian Holsteins. J Dairy Res 91(8):3184–3189. doi:10.3168/jds.2007-0947
Qiu H (2002) Modern dairy science. Agriculture Press of China, Beijing
Acknowledgments
This work was supported by the Key Development of New Transgenic Breeds Program of China (2009ZX08009-156B) and National Natural Science Foundation of China (31072016), and National “948” Project (2011-G2A), The authors would also like to thank the Dairy Association of China for providing frozen semen samples, DHI data and pedigree information. The authors also would like to thank Dr. David Rheinheimer, Post-doctoral Researcher, Wuhan University, for his assistance with English expression and editing.
Author information
Authors and Affiliations
Corresponding author
Additional information
M. A. Alim and Y. P. Fan are the authors have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Alim, M.A., Fan, Y.P., Wu, X.P. et al. Genetic effects of stearoyl-coenzyme A desaturase (SCD) polymorphism on milk production traits in the Chinese dairy population. Mol Biol Rep 39, 8733–8740 (2012). https://doi.org/10.1007/s11033-012-1733-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-1733-6