Abstract
Modern molecular interventions are dynamic gears for breeding animals with superior genetic make-up. These scientific efforts lead us toward sustainable dairy herds with improved milk production in terms of yield and quality. Many of candidate genes have been dissected at molecular level, and suitable genetic markers have been identified in cattle, but this work has not been validated in buffaloes so far. Stearoyl-coenzyme A desaturase (SCD) has been a potential candidate gene for fat content of milk. Genomic analysis of SCD revealed a total of six variations that were identified through DNA sequencing of animals with lower and higher butter fat %age. After statistical analysis, genotype AB of p.K158I could be associated (P value <0.0001) with higher milk fat %age (10.5 ± 0.5464). This SNP was validated on larger data set by cleaved amplified polymorphic sequences (CAPS) by using DdeI. To scrutinize the functional consequences of p.K158I, 3D protein structure of SCD was predicted by homology modeling and this variation was found located in the vicinity of functional domain and a part of transmembrane helix of this membrane integrated protein. This is a first report toward genetic screening of SCD gene at molecular level in buffalo. This report illustrates the implication of SCD gene and in particular p.K158I variation, in imparting its effect on milk fat %age, which can be targeted in selection of superior dairy buffaloes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Milk production is the area of focus for modern geneticists in recent years. Due to ever increasing demand of milk as nutritive dietary component, there are apprehensions regarding existing production potentials of dairy animal (both in yield and quality). So, recent scientific efforts are targeting genetic basis of milk production trait to identify genetic loci imparting significant role to enhance future production of dairy herds. Many of the candidate genes have been tested to find their role in determining milk production of animals (Jiang et al. 2010; Mullis 2007; Svennersten-Sjaunja and Olsson 2005; Li et al. 1990; Goddard and Hayes 2009; Grisart et al. 2002; Khatib et al. 2007, 2008). But, majority of identified markers have not been found informative in buffaloes, which are major dairy contributors in Asian regions and many other parts of world as well. So, there is need to work on genomic exploration of river buffalo for identification of novel selection signatures.
Stearoyl-coenzyme A desaturase (SCD) has been determined as potential gene effecting milk fatty acids profile (Gautier et al. 2006; Bauman et al. 2006; Soyeurt et al. 2006). SCD is a multifunctional complex enzyme important in the cellular biosynthesis of fatty acids (Ntambi and Miyazaki 2004). The SCD locus has been mapped on Bos taurus autosome (BTA)26 (Campbell et al. 2001). Some of significant associations have been reported in different cattle breeds (Ntambi and Miyazaki 2004; Campbell et al. 2001; Kgwatala et al. 2007; Taniguchi et al. 2004). The aforementioned results illustrate a probable significance of the SCD locus in gene-assisted selection programs for the genetic improvement of milk fat quality in dairy buffaloes as well.
In this context, present research was intended to screen the SCD gene in river buffalo of Pakistan. All six exons of the gene were sequenced and six variations were reported (Maryam et al. 2013). After statistical analysis, six variations were found nonsignificant potential loci (P > 0.05) for association studies. p.K158I was selected for obeying Hardy–Weinberg equilibrium (HWE) with significance (2.176 > 0.05). This region was genotyped by restriction digestion on larger buffalo population and was further subjected to association analysis. It was concluded that genotype AB was found associated with milk fat content in buffaloes.
This region was further analyzed for 3D protein structure to find functional consequences of the polymorphism. It was determined that p.K158I was located inside of the transmembrane region of SCD protein and in the vicinity of a functional domain. As this was a hydrophilic to hydrophobic substitution, this might have a role in increase of milk fat content in bovines.
Materials and methods
Phenotypic data set
River buffalo breed Nili-Ravi was selected as experimental animal because of its high milk butter fat potential (on an average 8 %). Animals were selected from Buffalo Research Institute (BRI), Pattoki, Pakistan, in their first month of second lactation. Fat %age (FP) of individual animal was taken from Production Record Data Sets available at farm. A total of 69 animals were selected for initial sequencing. On the basis of fat %age record, animals were categorized into two groups (group 1: FP > 8.0, group 2: FP < 8.0) and blood sampling was conducted.
Genomic DNA amplification and sequencing
Genomic DNA was extracted by using organic DNA extraction protocol reported by Maryam et al. (2012). Exonic region of SCD gene was amplified by using specific sets of primers reported by Alim et al. (2012). For initial study of gene, 69 animals (n = 34 from each group) were amplified by using 0.5 μl of each primer, 2.5 μl 109 PCR buffer, 2.5 mM each of dNTP, and 1 U of Taq DNA polymerase. The PCR protocol was 5 min at 94 °C for initial denaturing followed by 34 cycles at 94 °C for 30 s, 56 °C for 30 s, 72 °C for 30 s, and a final extension at 72 °C for 7 min for all the primers. Then, each PCR product was sequenced by Sanger’s chain termination method using the ABI3730XL (Applied Biosystems, Foster City, CA). Sequences from both groups were aligned on BLAST resource of NCBI and a total of six variations were identified (Table 1).
Statistical analysis
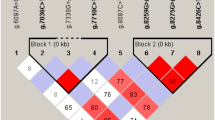
POPGENE (http://www.ualberta.ca/*fyeh/) version 1.32 was used to determine Hardy–Weinberg equilibrium (HWE) of each variation. Some of the variations could not be analyzed because of monomorphic nature in population. Some other was found significantly deviated from HWE and so could not be useful in association analysis. p.K158I was selected for further screening on larger data set (Fig. 1) and was further tested for probable association with milk fat % age.
CAPS
p.K158I was genotyped at larger data set to validate the association in animals from different livestock farms. Almost 346 more animals of Nili-Ravi buffaloes (Livestock Experimental Station, Bahadur nagar, Okara and Buffalo Colony, Karachi) were genotyped by cleaved amplified polymorphic sequences (CAPS). For this purpose, DdeI enzyme was selected and restriction was performed on PCR-amplified fragment of the SCD gene. Recipe for restriction digestion is given in Table 2. Restriction map was studied to determine the genotypic pictorial of the p.K158I (Fig. 2 and 3). After this screening, association analysis and genotypic frequencies were tested as mentioned in Tables 3, 4, and 5.
Association analysis
Association was performed by one-way ANOVA by calculating (mean ± SE). p.S80F was having weaker association with probability of 0.049444, while p.K158I was found to have stronger association with P ≤ 0.0001. Genotypic and allelic frequencies for these six loci were also calculated by POPGENE32 (Tables 3 and 4). It was found that genotype AB at p.K158I was strongly associated with high milk fat %age (10.5 ± 0.5464) (Table 5).
3D protein model prediction and analysis
Three-dimensional protein model was predicted by using PyMOL (www.pymol.org) and Phyre2 (www.sbg.bio.ic.ac.uk/phyre/html/). As no previous information could be retrieved for structural attributes of SCD protein, this model was further analyzed for prediction of probable functional domains, signal peptides, secondary structures, and transmembrane helix (Figs. 4 and 5).
Results
SCD gene was analyzed in buffaloes to identify genetic variations. A total of six variations were identified in exonic and some of intronic region of SCD. Exon-IV was found to have major proportion of all variations (31 %). This information is given in Table 1. After HWE testing (χ 2), SNPs obeying the Hardy–Weinberg law (Table 1) were further tested for association with fat content by calculating (mean ± SE). p.K158I was genotyped by CAPS on larger animal population and was found to have stronger association with higher fat content. Restriction mapping (Figs. 2 and 3) was done to validate the association probability at larger data set association of p.K158I illustrated that buffaloes with genotype AB were showing the maximum FP (P ≤ 0.0001) (Table 5). So, it was concluded that buffaloes with heterozygous genotype AG were depicting high FP than other genotypes.
Finally, 3D protein structure of SCD protein (359 amino acid residues) was predicted on the basis of sequence homology (Figs. 4 and 5). Predicted model for this protein was carrying 58 % alpha helices and 2 % beta sheets (Fig. 6). p.K158I was found as a part of transmembrane helix at position no. 158 (Table 6) and in the vicinity of steroid-binding domain. As this was part of functional protein and amino acid substitution was also hydrophilic to hydrophobic exchange, functional consequences of p.K158I could be predicted.
Discussion
A novel selection signature was identified in SCD gene that was found to be associated with higher milk fat content. A study was carried out by Macciotta et al. (2008), who claimed that cows with homozygous genotype CC at the SCD locus (g.10329C/T) producing more milk (about 2 kg/day) and protein (about 0.07 kg/day) compared with TT cows; heterozygous cows were in an intermediate position, but our results deviate from reported literature. The results of present research are contrasting than Macciotta but are in accordance with Alim et al. (2012) who also reported association of heterozygous genotypes with different milk production traits. B allele was found to have more frequency than A. As no efforts in terms of selective breeding have been initiated yet, so this might be due to natural genetic drift and provides a potential for future marker-assisted selection programs to enhance fat content of milk. Another study by Conte et al. (2006), on Italian brown cattle reported a significant association between SCD1 genotypes and milk FA C14:1 cis-9 and DI 14 (P B 0.01). In particular, the CC genotype was associated with 18.3 and 20.6 % more C14:1 cis-9 and DI 14, respectively, compared with the TT genotype. These apparently inconsistent results might be justified as experimental breed of this research was Nili-Ravi, which is a cross of Nili and Ravi buffalo breeds of Pakistan. This crossing although is being done over the past 100 years or even more, both breeds still hold a distinctive place among best dairy buffalo breeds in the world. So, cross-breeding of these animals resulted in pooling of better genotypes controlling different production traits. Now, because of its production potentials, better adaptability in the hot and humid climate of Pakistan, and better immunity, Nili-Ravi has been declared Black Gold of Asia.
Finally, 3D protein structure prediction was performed. Very limited literature information regarding protein structural attributes could be found. Stearoyl-CoA has no known paralogs. It, however, does have 19.4 % homologs in mammals and 20.8 % homologs in bacteria, so it is likely that these organisms have similar oxidoreductases that catalyze desaturation reactions as does this enzyme (Xiaoyun et al. 2013). The stearoyl-CoA desaturase system in animals is anchored in the endoplasmic reticulum membrane via four hydrophobic transmembrane domains with the active center (and the N- and C-termini) exposed to the cytosol (Buist 2004). Abovementioned literature reported transmembrane nature of SCD protein which is mitochondrial and is major enzyme in fatty acid synthesis. Two of the domains have been reported previously heme/steroid-binding domain and fatty acid desaturase domain (Buist 2004). p.K158I was found in the vicinity of steroid-binding domain, which is functionally active region for fatty acid synthesis. Observing, the abovementioned results following assumptions were made: This variation is switching of polarity to nonpolarity and hydrophilicity to hydrophobicity and so might have better transport across mitochondrial membrane and enhanced fatty acid production, or presence of this substitution in the vicinity of steroid-binding domain might affect protein functionality. These assumptions need further confirmations by advanced proteomic approaches. But, so far, this is first report regarding variation at amino acid level which is affecting fat content in the milk of bovines. These results suggest that SCD is a candidate gene that influences milk production traits and provides potentially useful information for dairy breeding programs.
References
Alim, M.A., Fan, Y. P., Wu, X. P., Xie, Y., Zhang, Y., Zhang, S. L., Sun, D. X., Zhang, Y., Zhang, Q., Liu, L. and Guo, G. 2012. Genetic effects of stearoyl-coenzyme A desaturase (SCD) polymorphism on milk production traits in the Chinese dairy population; Mol Biol Rep, 39,8733–8740
Bauman, D. E., Mather, I. H., Wall, R. J. and Lock, A. L. 2006. Major advances associated with the biosynthesis of milk; J. Dairy Sci, 89,1235–1243
Buist, P.H. 2004. Fatty acid desaturases: selecting the dehydrogenation channel; Nat. Prod. Rep, 21,249-262
Campbell, E. M. G., Callagher, D. S., Davis, S. K., Taylor, J. F. and Smith, S. B. 2001. Mapping of the bovine stearoyl-coenzyme A desaturase (SCD) gene to BTA26; J. Anim. Sci, 79,1954–1955
Conte, G., Mele, M., Castiglioni, B., Serra, A., Viva, M., Chessa, S., Pagnacco, G. and Secchiari, P. 2006. Relationship between bovine SCD polymorphism locus and mammary gland desaturation activity. In: 8th world congress on genetics applied to livestock production, Belo Horizonte, Minas Gerais, Brazil. Instituto Prociancia, pp: 22–37
Gautier, M., Barcelona, R. R., Fritz, S., Grohs, C., Druet, T., Boichard, D., Eggen, A. and Meuwissen, T. H. E. 2006. Fine mapping and physical characterization of two linked quantitative trait loci affecting milk fat yield on BTA26; Genetics, 172,425–436
Goddard, M.E. and Hayes, B.J. 2009. Mapping genes for complex traits in domestic animals and their use in breeding programmes; Nat Rev Genet, 10,381–391
Grisart, B., Coppieters, W., Farnir, F., Karim, L., Ford, C., Berzi, P., Cambisano, N., Mni, M., Reid, S. and Simon, P. 2002. Positional candidate cloning of a QTL in dairy cattle: identification of a mis-sense mutation in the bovine DGAT1 gene with major effect on milk yield and composition; Genome Res, 12(2),222–231
Jiang, L., Liu, J., Sun, D., Ma, P. and Ding, X. 2010. Genome wide association studies for milk production traits in Chinese Holstein population; PLoS ONE, 5(10), e13661
Kgwatala, P. M., Ibeagha-Awemu, E. M., Hayes, J. F. and Xhao, X. 2007. SNPs in the 3′ UTR of the stearoyl-CoA desaturase gene in Canadian Holstein and Jerseys; J. Dairy Sci, 90(Suppl. 1):420. (Abstr.)
Khatib, H., Zaitoun, I., Wiebelhaus-Finger, J., Chang, Y.M. and Rosa, G.J.M. 2007. The association of bovine PPARGC1A and OPN genes with milk composition in two independent Holstein cattle popu lations; J Dairy Sci, 90(6),2966–2970
Khatib, H., Monson, R.L., Schutzkus, V., Kohl, D.M., Rosa, G.J.M. and Rutledge, J.J. 2008. Mutations in the STAT5A gene are associated with embryonic survival and milk composition in cattle; J Dairy Sci, 91(2),784–793
Li, S., Crenshaw, E.B., Rawson, E.J., Simmons, D.M., Swanson, L.W. and Rosenfeld, M.G. 1990. Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene pit-1; Nature, 347, 528–533
Macciotta, N.P., Mele, M., Conte, G., Serra, A., Cassandro, M., Dal Zotto, R., Borlino, A.C., Pagnacco, G. and Secchiari, P. 2008. Association between a polymorphism at the stearoyl CoA desaturase locus and milk production traits in Italian Holsteins; J Dairy Res, 91(8),3184–3189
Maryam J., Babar, M.E., Nadeem, A. and Hussain, T. 2012. Genetic variants in interferon gamma (IFN-γ) gene are associated with resistance against ticks in Bos taurus and Bos indicus; Mol Biol Rep, 39(4),4565-70
Maryam, J., Nadeem, A. and Babar, M.E. 2013. Bovine stearoyl-CoA desaturase gene polymorphism in Nili Ravi Buffalo; Buffalo Bulletin, 32 (Special Issue 2), 710–713
Mullis, P.E. 2007. Genetics of growth hormone deficiency. Endocrinol Met Clinics North Ame, 36: 17–36
Ntambi, J. M. and Miyazaki, M. 2004. Regulation of stearoyl-CoA desaturases and role in metabolism; Prog. Lipid Res, 43,91–104
Soyeurt, H., Dardenne, P., Gillon, A., Croquet, C., Vanderick, S., Mayeres, P., Bertozzi, C. and Gengler, N. 2006. Variation in fatty acid contents of milk and milk fat within and across breeds; J. Dairy Sci, 89,4858–4865
Svennersten-Sjaunja, K. and Olsson, K. 2005. Endocrinology of milk production; Domest Anim Endocrinol, 29, 241–258
Taniguchi, M., Utsugi, T., Oyama, K., Mannen, H., Kobayashi, M., Tanabe, Y., Ogino, A. and Tsuji, S. 2004. Genotype of stearoyl CoA desaturase is associated with fatty acids composition in Japanese Black cattle; Mamm. Genome, 14,142–148
Xiaoyun, W.u., Xiaoju, Zou., Qing Chang, Yuru Zhang, Yunhai Li, Linqiang Zhang, Jingfei Huang, and Bin Liang. 2013. The evolutionary pattern and the regulation of stearoyl-CoA desaturase genes; BioMed Research International, Article ID 856521, 12 pages
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Maryam, J., Babar, M.E., Bao, Z. et al. A novel selection signature in stearoyl-coenzyme A desaturase (SCD) gene for enhanced milk fat content in Bubalus bubalis . Trop Anim Health Prod 48, 1343–1349 (2016). https://doi.org/10.1007/s11250-016-1092-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11250-016-1092-8