Abstract
C2H2 proteins belong to a group of transcription factors (TFs) existing as a superfamily that plays important roles in defense responses and various other physiological processes in plants. The present study aimed to screen for and identify C2H2 proteins associated with defense responses to abiotic and biotic stresses in Carica papaya L. Data were collected for 47,483 papaya-expressed sequence tags (ESTs). The full-length cDNA nucleotide sequences of 87 C2H2 proteins were predicated by BioEdit. All 91 C2H2 proteins were aligned, and a phylogenetic tree was constructed using DNAman. The expression levels of 42 C2H2 were analyzed under conditions of salt stress by quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR). Methyl jasmonate treatment rapidly upregulated ZF 23.4 and ZF 30,912.1 by 18.6- and 21.7-fold, respectively. ZF 1.3, ZF 138.44, ZF 94.49, ZF29.160, and ZF 20.206 were found to be downregulated after low temperature treatment at very significant levels (p < 0.01). ZF 23.4, ZF 161.1, and ZF 30,912.1 were upregulated while ZF1.3, ZF 158.1, ZF 249.5, ZF 138.44, ZF 94.49, ZF 29.160, and ZF20.206 were significantly downregulated by Spermine treatment. ZF 23.4 was upregulated while ZF 1.3, ZF 249.5, ZF94.94, ZF 29.160, ZF 138.44, and ZF 20.206 were significantly repressed after SA treatment. ZF 23.4 and ZF 30,912.1 were significantly upregulated after sap inoculation with papaya ringspot virus pathogen. ZF 30,912.1 was subcellularly localized in the nucleus by a transgenic fusion of pBS–ZF 30,912.1–GFP into the protoplast of papaya. The results of the present study showed that ZF 30,912.1 could be an important TF that mediates responses to abiotic and biotic stresses in papaya.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Carica papaya L. is an important fruit in tropical and subtropical countries. Severe losses of production may occur in Carica papaya L. under conditions of abiotic stressors, such as extremely low (<−2°C) or high temperature (>35°C), waterlogging, drought, and salt stress caused by excessive use of chemical fertilizers, and biotic stresses, such as papaya ringspot virus (PRSV) infection. Thus, efforts are continuously being made to develop papaya varieties that have better tolerance to these stresses.
The response of plants to abiotic and biotic stresses is more likely to be a polygenic cooperative defense response induced by cold, drought, salt, wounding, and disease rather than the single response of a single gene [1]. Therefore, in improving the stress tolerance of plants through molecular breeding, the traditional transformative method that applies a single resistance gene has limitations. Transcription factors (TFs) that regulate the expression of multiple genes are more promising candidates for manipulation [2]. TFs regulate gene expression in response to various external and internal cues by activating or suppressing downstream genes in the pathway. Zinc finger proteins (ZFPs) are a group of TFs that are expressed in different stressful conditions. Previous studies have indicated that some C2H2 ZFPs are induced by various stresses and may be important candidate genes. For example, the ZFP182 is markedly induced in paddy seedlings by cold, salt, and ABA treatments. The expression of ZFP182 in transgenic tobacco and overexpression in rice increases plant tolerance to salt stress [3]. C2H2 ZFP and pathogenic StZFP1 of potato are constitutively expressed in different organs of mature plants when subjected to salt, dehydration, and exogenously applied ABA. StZFP1 expression also occurs in response to infection by the late blight pathogen Phytophthora infestans, and the ectopic expression of StZFP1 increases plant tolerance to salt stress in transgenic tobacco [4].
C2H2 ZFPs constitute one of the largest TF families in plants [5], and their presence has been reported in several plant species, including Arabidopsis, petunia, rice, wheat, soybean, and pepper. In Arabidopsis, a total of 176 C2H2 ZFPs have been uncovered, constituting the abundant family of putative transcriptional regulators [6]. The genome of Oryza sativa codes for 189 C2H2 ZFP TFs [7]. The C2H2 protein is a major TF that regulates plant responses to various stresses and plant development [3, 4, 8]. In 2008, Ming and colleagues [9] compared gene numbers between the TF tribe and related tribes of Arabidopsis and papaya and reported that most TFs were represented by fewer genes in papaya than in Arabidopsis.
Previous studies have reported that C2H2 plays a very important role in the defense responses of many plants. C2H2 has been found to enhance the tolerance of plants to salt stress, some examples of which include TFIIIA-type ZFP with streptozotocin (STZ) [10], AtNAC2 [11], the EAR-motif of C2H2 in Arabidopsis [12], a R1R2R3-type myeloblastosis (MYB) gene [13], ZFP179 [8], and ZFP DST in rice [14, 15]. ZFPs contribute significantly to the temperature stress tolerance of certain varieties of paddy [16–18], Arabidopsis [19], cotton [20], and soybean [21]. However, the role of papaya C2H2 in mediating defense responses to stresses has not been reported thus far.
Salicylic acid (SA) and methyl jasmonate (MeJA) are essential signal molecules involved in many aspects of plant development, as well as the responses of plants to biotic stresses, especially to pathogen infection, wounding, and insect attack [22–25]. SA plays a positive role in plant defenses against biotrophic pathogens, whereas jasmonic acid/ethylene (JA/ET) appears to be important in the defense against necrotrophic pathogens [26]. Early studies have indicated that polyamine is involved in plant responses to different environmental stresses. In addition, there is genetic potential for modulation of the polyamine levels of plants to cope with abiotic stress conditions [27–29].
Identification and evaluation of C2H2 related to stress tolerance are essential for molecular enhancements in papaya breeding. The present study aimed to identify prominent C2H2 that evince rapid responses to stressors, such as Spm, SA, MeJA, salt, low temperature, and PRSV infection. Data of the papaya C2H2 superfamily were collected using an ExPasy proteomics server (http://cn.expasy.org), a BLAST local database of expressed sequences tags (ESTs) of papaya was built, and the exact nucleotide sequence of C2H2 in the papaya superfamily were obtained using Bioedit software. The phylogenetic tree of C2H2 was constructed using DNAman software. Real-time PCR was used to quantitate the expression level of ZF in response to stressors and determine TF that are potential candidates of interest for improving stress tolerance through transgenic modification.
Materials and methods
Plant materials and treatments
Seeds of Carica papaya L. ‘Hawaii’ were obtained from Shanya City in Hainan Province, China. Sprouts were grown in sterile vermiculite and incubated in illumination incubators at 28°C under a photoperiod of 14 h/d. Treatments were carried out on 35-day-old seedlings with 5–6 leaves. In preliminary experiments, several concentrations of spermine (Spm), salt (NaCl) solution, SA, and MeJA were separately tested, and the highest possible concentration for each stressor that did not induce necrosis was selected. Each treatment involved the leaves of five plants, and samples were taken from experiments conducted in triplicate. Plants treated only with water were used as controls.
Different groups based on treatments
In Group A, 200 mmol/L salt solution (10 mL/plant) was carefully dropped from the apical point of the plants and allowed to flow naturally, aided by gravity. Group B comprised tests where 0.5 mmol/L Spm was carefully sprayed over two cotyledons and two euphilla at a dose of 10 mL/plant. In Group C, 1 mmol/L SA was sprayed onto the cotyledons and two euphilla at a dose of 10 mL/plant. Group D experiments involved distribution of droplets of 1 mmol/L MeJA at a dose of 1 mL/plant. Group E plants were kept at 4°C for 1 h in the dark, and then moved to the growth incubator at a temperature of (25 ± 1)°C; samples were collected at different intervals. Plants with and without treatment were placed in different growth chambers. The euphylla were harvested and frozen immediately in liquid nitrogen and stored at −70°C until RNA extraction. Basal levels of C2H2 expression were assessed in leaves, stems, and roots of papaya. The C2H2 expression levels recorded were normalized against ACTIN transcript accumulation. The PRSV pathogen was inoculated and samples were collected after 24 h.
Database searches and gene annotation
Through superfamily 1.75 (HMM library and genome assignments server; http://supfam.org), the protein sequences corresponding to each papaya C2H2 unigene were predicated. The specific ZF C2H2-type domain signature and QALGGH motif were investigated by searching the ExPasy proteomics server (http://cn.expasy.org). Weak hits were subsequently excluded. The C2H2 of papaya were aligned and a phylogenetic tree was constructed by using DNAman software (gap opening 10, gap extension 2, Blosum weight matrix). A BLAST local database of the papaya ESTs was constructed with BioEdit software using several EST sequences (EX227656–EX303501) for comparison, and the nucleotide sequence of C2H2 ZF were confirmed. The EAR motifs in ZFP proteins were identified according to known EAR-motifs and the DLN-box [3, 30, 31].
RNA extraction and qRT-PCR
Total RNA was extracted from papaya leaf, root, and stem samples using TRIzol reagent (Invitrogen) and subjected to DNase I digestion for purification. RNA samples were detected using an Ultrospec2100 UV/Visible spectrophotometer (Amershan GE Healthcare, USA), and 1% agarose gel was run to visualize the quality of the RNA. First-strand cDNA was synthesized from 2 μg of total RNA at a final volume of 20 μL using TransScript first-strand cDNA synthesis superMix (Transgen) and oligo-dT(18) primers according to the manufacturer’s instructions.
A qRT-PCR assay of gene expression-specific primers was designed from the papaya cDNA sequences using Primer Express 3.0 software, with temperatures in the range of 58–60°C, primers lengths of 19–24 bp, and amplification fragment lengths measuring 150–200 bp. Where possible, primers were designed outside the conserved ZF C2H2-type domain to ensure gene-specific primers; primer pairs are listed in Supplementary Table 1. The primers were synthesized by Sanggon Shanghai Biology Technology Ltd., at a final concentration of 0.2 μmol/L. qRT-PCR was performed using a Line Gene K system (Bioer Technology, Japan), and the software program for comparative quantitation PCR was run for 1.3 ng cDNA in a 20 μL amplification mixture containing SYBR Green (Invitrogen). The cycling conditions are as follows: 2 min at 50°C, followed by 3 min polymerase activity at 95°C and 35 cycles at 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s. Each assay was conducted in triplicate and included a no-template control. The threshold cycle (Ct) of the primary amplification curve was used for calculations. The ACTIN gene was selected as the internal constitutively expressed control (normalization), according to the following formula:
Data were analyzed using the 2−∆∆Ct method [32]. In general, genes whose expressions changed more than twofold in response to stress treatment were specifically noted. Dilutions of cDNA (1:10–1:1,000) from a reference sample were used to construct a relative standard curve. For all primer pairs tested, efficiencies varied from 0.9 to 1.10 (data not shown). The specificity of PCR results was checked by melting-curve analyses, and only primer sets producing a single sequence-specific peak were conserved. Data were analyzed using the FQDpcr software package to obtain relative expression levels of the papaya gene based on the comparative Ct method; data are either expressed as fold changes relative to the calibrator average or as log (base 2)-fold changes relative to the calibrator average. Data were analyzed using Microsoft Excel.
Subcelluar localization of the ZF30912.1-GFP fusion protein
RNA was used as the reverse transcription element for the ZF 30,912.1 and cDNA was the template. The gene fragment was cloned, recovered, and constructed into the pMD-T19 vector and confirmed by sequencing. The forward primer was 5′-ATGAAGAGAAGTAATTTG-3′ and the reverse primer was 5′-GAAGAAGCAATCCAC-3′. To confirm that the ZF30,912.1 protein was indeed the target in the nucleus, the ZF 30,912.1 was selected for analysis in vivo. The coding regions of ZF 30,912.1 were fused to the N-terminus of synthetic green fluorescent protein (GFP) and expressed in papaya under the control of the constitutive cauliflower mosaic virus 35S promoter. The fusion protein-expression vector pBS–ZF 30,912.1–GFP was constructed. Papaya protoplasts were isolated and either the pBS–GFP or the pBS–ZF 30,912.1–GFP plasmid was transformed using the PEG method. Previous reports have proven that pBS–GFP is expressed in the nucleus and cytomembrane [33, 34]. GFP expression and subcellular localization were observed using a confocal laser scanning microscope (LSM 510 META; Zeiss, Germany) with excitation and emission wavelengths of 475 and 520 nm, respectively (pixel depth: 12 bit, objective, plan-A; Pochromat 63×/1.4 oil DIC), and imaged using the associated software (Carl Zeiss Image Microscopy Release 4.0).
Results
Identification of C2H2 transcription factor
To assess the characteristics of the C2H2 superfamily proteins in Carica papaya, data were collected on a set of 91 expressed sequences of protein. Data of 47,483 papaya ESTs were collected and a BLAST local database was built with the BioEdit software. A total of 87 C2H2 were identified, and their full-length nucleic acid sequences were obtained from six reading frames using the tblastn process, according to the percentage of homology, GenBank accession number, and corresponding position of nucleotide sequence; details of these 87 C2H2 are shown in Supplementary Data Tables 2-1, 2-1, and 2-3.
Conserved ZF C2H2-type domain signatures and motifs were analyzed. Supplementary Table 2 shows 91 amino acid sequences with the typical family structure, including the diagnostic signatures of C-X2, 4-C-X3-Φ-X5-Φ-X2-H-X3, 4-H, and with the corresponding genes encoding 1–5 C2H2 domains. Among these, 38 ZFPs had a ZF QALGGH motif, five ZFPs had a two-ZF QALGGH motif, and one ZFP had a three-ZF QALGGH motif. 11 C2H2 proteins possessed the HKH motif–containing C2H2 ZF, three C2H2 proteins comprised a BED-type ZF, and 39 contained the classic ZF motif, of which six contained two types of ZF. Among the 91 ZFP subfamily members, 15.4% contained an EAR-motif while 17.6% contained nucleus location signatures (DLN-box). The protein name, size (amino acids), GenBank accession number, frame, corresponding position of available nucleotide sequences, location of C2H2 domain signature, EAR motif, and DLN-box of each protein are listed in supplementary data Tables 2-1, 2-1, and 2-3.
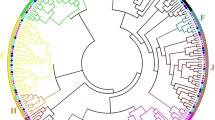
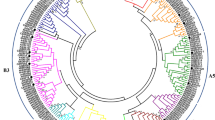
Phylogenetic analysis of C2H2 proteins
An unrooted tree is shown for 91 ZF motifs to evolutionary relationships among different types of C2H2 ZF motifs of papaya. The number of ZFPs in the proteins of this superfamily varied from one to five domain signature(s): ϕ-X-C-X2,4-C-X3-ϕ-X5-ϕ-X2-H-X3,4-H. The C2H2 domain sequences of the papaya proteins were aligned. An unrooted plot tree was also made, exclusively comprising all ZF domains from the first cysteine to the last histidine. In this unrooted plotted phylogenetic tree comparing the C2H2 proteins of papaya, proteins containing classic ZF motifs are shown in Supplementary Fig. 1a, d. Q-type C2H2 proteins with the QALGGH motif were arranged together in order, as shown in Supplementary Fig. 1b. Proteins contained two classic ZFs (Supplementary Fig. 1c); those containing one or more classic ZFs, or those with one or more HKH motifs, are shown in Supplementary Fig. 1e.
C2H2 TFs are involved in the early response of papaya to salt stress
The effects of salt stress were determined relative to ACTIN housekeeping genes in leaves 12, 24, and 48 h after treatment of 42 ZF analyzed genes with 200 mmol/L salt (Fig. 1a). According to the t-test analysis of the average value of the samples and control in pairs, the expression levels of ZF 9.152, ZF 30,912.1, ZF 48.203, ZF 92.92, ZF 190.29, and ZF 1.3 were downregulated after 12 h of salt stress treatment at very significant levels (p < 0.01), those of ZF 30.114, ZF 9.152, ZF 48.203, ZF 92.92, ZF 190.29, and ZF 1.3 were downregulated after 24 h of salt stress treatment at very significant levels (p < 0.01), those of ZF 30,912.1 and ZF 45.58 decreased at significant levels (p < 0.05), and those of ZF 9.152, ZF 30,912.1, ZF 45.58, ZF 92.92, ZF 158.1, and ZF 190.29 were downregulated after 48 h at very significant levels (p < 0.01). The salt solution significantly activated the expressions of ZF 29.160, ZF 20.206, ZF 9.185, ZF 138.44, and ZF 169.30 after 24 h and decreased thereafter, while the expression levels of ZF 28.115 and ZF 4.50 were upregulated very significantly after 48 h of the same treatment (Fig. 1c). The expression levels of other ZF were changed but not significantly, as shown in Fig. 1a–c.
Gene expression analysis by quantitative RT-PCR for the salt stress test. The code of evm.TU.supercontig C2H2 transcription factors is shown in the x axis. Expression levels were calculated relative to ACTIN expression. Expression levels are presented as fold-induction relative to the control. Mean ± SE of triple experiments are shown. a Expression levels of all 22 ZFs were relatively downregulated compared with ACTIN and calculated by the formula 2−∆∆Ct. b Different ZFs were upregulated and downregulated by salt treatment. Expression levels of ZFs increased after salt stress treatment
C2H2 ZFs are involved in early response to low temperature in papaya
After subjecting papaya plants to low temperature, leaves were harvested after 1, 4, and 24 h. The expression level of ZF 23.4 was significantly induced after 24 h (p < 0.05) while that of ZF 161.1 was induced at 4 and 24 h at significant levels (p < 0.05). The expressions of ZF 1.3, ZF 94.49, and ZF 20.206 were found to be downregulated at 1 h, those of ZF 1.3, ZF 138.44, ZF29.160, and ZF 20.206 were found to be downregulated at 4 h, and those of ZF 1.3, ZF 94.49, and ZF29.160 were found to be downregulated at 24 h; all difference analysis results showed significant levels of variation (p < 0.01) (Fig. 2).
C2H2 ZFs are involved in early response of papaya to spermine
Spm plays an essential role in the response of plants to abiotic stress. After Spm treatment, ZF 23.4 was upregulated by 2.3- and 4.4-fold after 4 and 12 h, respectively, ZF 161.1 was upregulated by 2.8-fold after 4 h at very significant levels (p < 0.01), and ZF 30,912.1 was upregulated by more than twofold after 4 h at significant levels (p < 0.05). The expression levels of ZF 1.3, ZF 138.44, ZF 94.49, ZF 29.160, and ZF20.206 were downregulated at very significant levels (p < 0.01) after 12 h. ZF 1.3, ZF 158.1, ZF 249.5, ZF 138.44, ZF 94.49, ZF 29.160, and ZF 20.206 were also downregulated at very significant levels (p < 0.01) after 24 h (Fig. 3).
The expression pattern of 15 ZF in papaya treated with and without Spm. The first column represents the expression levels of ZF compared with those of ACTIN without Spm treatment as the control. The three other columns represent the expression levels of ZF in Spm-treated plants. ZF23.4, ZF161.1, and ZF30912.1 were upregulated by more than twofold compared with the control
C2H2 ZFs are involved in the early response to SA
The expression patterns of ZFs were examined in response to SA. The ZF 23.4 was upregulated 2.4-fold at 4 h after SA treatment but decreased thereafter. The expression levels of ZF 23.4 were upregulated at 4, 12, and 24 h at very significant levels (p < 0.01) compared with the control (Fig. 4). ZF 92.92, ZF 1.3, ZF 138.44, ZF 29.160, and ZF 20.206 were rapidly repressed after 4 h at very significant levels (p < 0.01), ZF 1.3, ZF 249.5, ZF94.94, ZF 29.160, and ZF 20.206 were rapidly repressed after 12 h at very significant levels (p < 0.01), and ZF 1.3, ZF 249.5, ZF 138.44, ZF 94.94, ZF 29.160, and ZF 20.206 were rapidly repressed after 24 h at very significant levels (p < 0.01).
ZF 23.4 and ZF 30,912.1 are involved in the early response of papaya to MeJA
To further characterize the expression patterns of ZF in response to MeJA, a time course expression pattern was determined by qRT-PCR. The ZF displayed different induction patterns when subjected to MeJA treatment. As shown in Fig. 5, of the 15 genes analyzed, the expression of two genes were upregulated prominently: ZF 23.4 was rapidly induced by 18.6-, 8.5-, and 6.7-fold while ZF 30912.1 was rapidly induced by 9.3-, 21.7-, and 21.4-fold after 4, 12, and 24 h of MeJA treatment, respectively. Peak expression levels of these genes occurred after 4 and 12 h, respectively. The two genes showed exceptionally sensitive responses to MeJA treatment at very significant levels (p < 0.01). The expression level of ZF 161.1 was induced by 2.4-, 2.1-, and 4-fold after 4, 12, and 24 h, respectively. Otherwise, the expression level of ZF 1.3 was downregulated after 4, 12, and 24 h at very significant levels (p < 0.01). Furthermore, the expression level of ZF 138.44 was decreased after 12 h at very significant levels (p < 0.01) and the expression levels of ZF 29.160 and ZF 20.206 were downregulated after 12 and 24 h at very significant levels (p < 0.01).
C2H2 expression in different plant organs and leaves inoculated with PRSV pathogen
Basal expression levels of leaves, stems, and roots varied for different genes. The mRNA expression pattern of six C2H2 ZF in different organs was further characterized in this study. The expression level of ZF 92.92 in the stem was lower by 0.03-fold, corresponding to the expression of the ACTIN gene. The experiment revealed stem tissue-specific accumulation. The expression levels of ZF 23.4 and ZF 30,912.1 were induced by 2.4- and 2.1-fold 24 h after sap inoculation with PRSV pathogen at significant levels (p < 0.05) (Fig. 6).
Sequencing of ZF 30,912.1 and localization of ZF30,912.1 C2H2 proteins
After 600 bp fragment amplification, the ZF30,912.1 was cloned and constructed into the pMD-T19 vector. Sequencing and analysis indicated that the 600 bp sequence shared 100% homology with the sequence from GenBank, gi|186136405|, frame (−1), for the full length from 2116 to 1517 bp (Supplementary Table 2; Fig. 2a). The amino acid sequence of the C2H2-type ZFP30,912.1 were compared with ZF domains of 12 proteins, which involved in environmental stress and organ development. The results are shown in Fig. 2b. ZFP30,912.1 was homologous to CAZFP1, SCOF1, AZF1, BCZFP1, ZPT2-1, ZPT2-2, AZF3, OSZFP34, ZPT3-3, ZPT2-10, and ZPT3-2 by 61.9, 61.9, 57.2, 57.1, 37.7, 61.9 66.7, 61.9, 76.2, 76.2, and 57.1%, respectively.
Analysis of the ZFP30,912.1 amino acid sequence indicated that it contained two canonical Cys2/His2 ZF motifs comprising 21 amino acids and separated by 39 amino acids. In addition to these two well-conserved motifs, three other more or less complete regions were observed: a hydrophobic leucine-rich region (L-box), probably involved in protein–protein interaction, a C-terminal region DLN-box, and a residue conserved among all 13 sequences (Supplementary Fig. 2b).
Transient expression analysis of the ZF30,912.1-GFP fusion protein revealed that ZFP30,912.1 was localized preferentially in the nucleus. Subcellular localization of the C2H2 ZF 30,912.1 was examined using GFP. The control plasmid pBS–GFP and the GFP fusion gene pBS–ZF 30,912.1–GFP were separately transformed into the protoplasts of papaya. Under a confocal laser scanning microscope, the control plasmid pBS–GFP was observed to express GFP in the nucleus and cytomembrane (Fig. 7a–c), whereas the GFP fusion protein gene of pBS-ZF 30912.1-GFP was transformed and expressed only in the nucleus (Fig. 7d–f).
Nuclear localization of the ZF30,912.1 protein in the protoplast of papaya in 25-days-old leaves. In the present study, 25-days-old leaves were observed by confocal microscopy. a–c The control vector pBS-GFP was transformed. d–f The subcellular localization of pBS–ZF30912.1–GFP. In each case, the left panel shows GFP fluorescence, the middle panel shows red fluorescence, and the right panel represents overlays of bright-field and fluorescence images
Discussion
C2H2 ZFs have been reported to regulate gene expression under biotic and abiotic stresses in several plant species [35, 36]. C2H2 ZFPs in papaya, however, have not been functionally characterized thus far. The present study is the first to provide data of the induced expression of C2H2 ZF TFs under biotic and abiotic stresses in papaya. Nucleotide sequence information of C2H2 was obtained using a conventional bioinformatics method. Conserved ZF C2H2-type domain signatures and motif characteristics were described, and the phylogenetic relationships among 91 major members were analyzed. Expression levels of some C2H2 ZF in response to abiotic stresses, such as salt and cold (4°C), and biotic stresses or their signal molecules, such as Spm, SA, and MeJA, as well as PRSV infection, were investigated. The present study provides information that may help facilitate cloning and further functional studies of C2H2 in papaya.
Abiotic stresses involve the negative impacts of non-living factors on living organisms that occur within a specific environment, while biotic stresses are caused by living organisms, such as fungi, bacteria, viruses, and nematodes, among others. In the present experiment, two common abiotic stresses, salt and cold (4°C), were artificially induced, and SA, MeJA, and Spm were used as biotic signal molecules, in addition to PRSV infection, to study abiotic and biotic stresses. ZFP30,912.1 was transiently upregulated by MeJA, Spm, and PRSV infection by 21.7-, 3.8-, and 2.1-fold, respectively. ZFP 23.4 expression levels were enhanced by 18.6-, 2.4-, 4.4-, 2.4-, and 1.7-fold after MeJA, SA, Spm, PRSV, and cold treatments. However, salt stress downregulated the expressions of FP 23.4 and ZF 30,912.1 by 0.01- and 0.08-fold, respectively. ZFP 30,912.1 and ZFP 23.4 were considered the top-priority genes for function validation.
Studies have previously reported that all C2H2 responses to stresses in plants have a double ZF structure [37, 38]. ZFP30,912.1, a typical ZFP, has two C2H2 ZF motifs, each having a conserved QALGGH sequence; this QALGGH motif has so far been reported only in plants [5]. However, ZF 23.4 contains three classic ZFs and no QALGGH motifs or EAR-box. It is possible that the ZF 23.4, and ZF 30,912.1 C2H2 play diverse roles in modulating defense responses against multiple biotic and abiotic stresses. Further, ZF 161.1 comprises four classic ZFs, the expression levels of which were upregulated by 2.8- and 4-fold after either Spm or MeJA biotic treatment. ZF 1.3, ZF 138.44, ZF 29.160, and ZF 20.206 contained a QALGGH sequence and were downregulated very significantly by cold stress; thus, these genes are likely related to cold stress. The results of the present study indicate that the responses of C2H2 to stresses in plants are not restricted to the double ZF structure.
Researchers have previously reported that TF genes function in various ways to confer stress tolerance to plants [39–41]. SA and MeJA are key signal molecules involved in plant disease-defense responses [42]. ZF 23.4 and ZF 30,912.1 were strongly induced by 18.6- and 21.7-fold after MeJA treatment, while the ZF 23.4 was induced by 2.4-fold and ZF 30,912.1 was induced by less than twofold after SA treatment. These findings indicate that MeJA is a more sensitive signal molecule than SA for ZF 23.4 and ZF 30,912.1 expression under stress.
A previous report demonstrated that the EAR domain of a C2H2 ZFP plays an important role in the defense response of Arabidopsis to abiotic stress [12]. In the present experiment, all ZFPs containing the DLN-box/EAR motif, including ZF 6.32, ZF 14.159, ZF 20.206, ZF 53.160, ZF 92.92, ZF 138.44, ZF 163.28, ZF 190.29, ZF 249.5, ZF 30,912.1, ZF 2027.1, ZF 33,076.1, and ZF 1.3, were distributed in Group B of the phylogenetic unrooted tree; only ZF 1.103 was located in Group E. ZF 29.160 and ZF 12.70 only contained the DLN-box and were located in Group B. All ZFs in Group B belong to Q-type C2H2 proteins with the QALGGH motif. The ZFPs analyzed were distributed in the B, C, D, and E positions of the phylogenetic tree relative to one or more stress conditions, as shown in Supplement Fig. 1. Further functional validation of these genes may help identify candidate genes that could contribute to the defense responses of transgenic papaya plants.
Proteins with similar domains may have identical or similar biologic functions [43]. The ZF 30,912.1 was cloned and subcellular localization assay indicated that ZFP30,912.1 localizes in the nucleus. These results prove that ZFP 30,912.1 may function in the nucleus. The results of organic expression analysis indicated that ZFP 30,912.1 is constitutively expressed in roots, stems, and leaves of papaya. The sequence of ZF 30,912.1 was compared with those of 12 ZFPs with known functions, including improvements in disease resistance and drought, salt, and cold tolerance, as well as a number of development-related events. ZF 30,912.1 shares homologies with 12 ZFPs of between 37.7 and 76.2%. Whether or not ZF 30,912.1 possesses functions identical or similar to these 12 ZFPs can be verified by gene transformation and gene silencing techniques, such as RNAi and VIGS. The data generated in the present experiments will facilitate further functional genomics studies to understand the precise role of C2H2 in modulating stress responses in papaya and other plants.
Abbreviations
- ABA:
-
Abscisic acid
- SA:
-
Salicylic acid
- C2H2:
-
Cys2/His2 finger protein
- EAR:
-
ERF associated amphiphilic repression
- GFP:
-
Green fluorescent protein
- MeJA:
-
Methyl-jasmonate
- PEG:
-
Polyethylene glycol
- RNAi:
-
RNA interference
- Spm:
-
Spermine
- TF:
-
Transcription factor
- TFPs:
-
Transcription factor proteins
- VIGS:
-
Virus induced gene silencing
- ZF:
-
Zinc finger
- ZFP:
-
Zinc finger protein
- qRT-PCR:
-
Real-time quantitative PCR
References
Zhang James Z, Creelman Robert A, Zhu Jian-Kang (2004) From laboratory to field. Using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops. Plant Physiol 135:615–621
Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9:436–442
Huang J, Yang X, Wang MM, Tang HJ, Ding LY, Shen Y, Zhang HS (2007) A novel rice C2H2-type zinc finger protein lacking DLN-box/EAR-motif plays a role in salt tolerance. Biochim Biophys Acta 1769:220–227
Tian ZD, Zhang Y, Liu J, Xie CH (2010) Novel potato C2H2-type zinc finger protein gene, StZFP1, which responds to biotic and abiotic stress, plays a role in salt tolerance. Plant Biol (Stuttg) 12(5):689–697
Kubo K, Sakamoto A, Kobayashi A, Rybka Z, Kanno Y, Nakagawa H, Takatsuji H (1998) Cys2/His2 zinc-finger protein family of petunia: evolution and general mechanism of target-sequence recognition. Nucleic Acids Res 26:608–615
Englbrcht CC, Schoof H, Bohm S (2004) Conservation diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics 5:39
Agarwal P, Arora R, Ray S, Singh AK, Singh VP, Takatsuji H, Kapoor S, Tyagi AK (2007) Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol Biol 65:467–485
Sun SJ, Guo SQ, Yang X, Bao YM, Tang HJ, Sun H, Huang J, Zhang HS (2010) Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J Exp Bot 61(10):2807–2818
Ming R, Hou S, Feng Y et al (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452:991–996
Xu SM, Wang XC, Chen J (2007) Zinc finger protein 1 (ThZF1) from salt cress (Thellungiella halophila) is a Cys-2/His-2-type transcription factor involved in drought and salt stress. Plant Cell Rep 26:497–506
He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44:903–916
Cifrci-Yilmaz S, Morsy MR, Song L, Coutu A, Krizek BA, Lewis MW, Warren D, Cushman J, Connolly EL, Mittler R (2007) The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J Biol Chem 282:9260–9268
Dai X, Xu Y, Ma Q, Xu W, Wang T, Xue Y, Chong K (2007) Overexpression of a R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol 143:1739–1751
Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX (2009) A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev 23:1805–1817
Oh S, Song S, Kim Y, Jang H, Kim S, Kim M, Kim Y, Nahm B, Kim J (2005) Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol 138:341–351
Li G, Tai FJ, Zheng Y, Luo J, Gong SY, Zhang ZT, Li XB (2009) Increased tolerance of rice to cold, drought and oxidative stresses mediated by the overexpression of a gene that encodes the zinc finger protein ZFP245. Biochem Biophys Res Commun 389(3):556–561
Cuevas JC, Lopez-Cobollo R, Alcazar R, Zarza X, Koncz C, Altabell T, Salinas J, Tiburcio AF, Ferrando A (2009) Putrescine as a signal to modulate the indispensable ABA increase under cold stress. Plant Signal Behav 4:219–220
Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 47:141–153
Vogel JT, Zarka DG, Buskirk VHA, Fowler SG, Thomashow MF (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41(2):195–211
Kim JC, Lee SH, Cheng YH, Yoo CM, Lee SI, Chun HJ, Yun DJ, Hong JC, Lee SY, Lim CO, Cho MJ (2001) A novel cold inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plant. Plant J 25:247–259
Li G, Tai FJ, Zheng Y, Luo J, Gong SY, Zhang ZT, Li XB (2010) Two cotton Cys2/His2-type zinc-finger proteins, GhDi19-1 and GhDi19-2, are involved in plant response to salt/drought stress and abscisic acid signaling. Plant Mol Biol 74:437–452
Lorenzo O, Solano R (2005) Molecular players regulating the jasmonate signalling network. Curr Opin Plant Biol 8:532–540
Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16:1938–1950
Koornneef A, Pieterse CM (2008) Cross talk in defense signaling. Plant Physiol 146:839–844
Wang Z, Cao GY, Wang XL, Miao J, Liu XT, Chen ZL, Qu LJ, Gu HY (2008) Identification and characterization of COI1-dependent transcription factor genes involved in JA-mediated response to wounding in Arabidopsis plants. Plant Cell Rep 27:125–135
Yang B, Jiang YQ, Rahman MH, Deyholos MK, Kav NN (2009) Identification and expression analysis of WRKY transcription factor genes in canola (Brassica napus L.) in response to fungal pathogens and hormone treatments. BMC Plant Biol 9:68
Groppa MD, Benavides MP (2008) Polyamines and abiotic stress: recent advances. Amino Acids 34:35–45
Alonso-Blanco C, Aarts MGM, Bentsink L, Keurentjes JJB, Reymond M, Vreugdenhil D, Koornnee M (2009) What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21(7):1877–1896. www.plantcell.org/cgi/doi/10.1105/tpc.109.068114
Gill SS, Tuteja N (2010) Polyamines and abiotic stress tolerance in plants. Plant Signal Behav 7:5(1)
Kam J, Gresshoff PM, Shorter R, Xue GP (2008) The Q-type C2H2 zinc finger subfamily of transcription factors in Triticum aestivum is predominantly expressed in roots and enriched with members containing an EAR repressor motif and responsive to drought stress. Plant Mol Biol 67:305–322
Wang GF, Wang H, Zhu J, Zhang J, Zhang XW, Wang F, Tang YP, Mei B, Xu ZK, Song RT (2010) An expression analysis of 57 transcription factors derived from ESTs of developing seeds in Maize (Zea mays). Plant Cell Rep 29:545–559
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real time quantitative PCR and the 2−∆∆Ct method. Methods 25:402–408
Yoo Sang-Dong, Cho Young-Hee, Sheen Jen (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2(7):1565–1572
Jiang L, Wang J, Liu Z, Wang L, Zhang F, Liu GC, Zhong Q (2010) Silencing induced by inverted repeat constructs in protoplasts of Nicotiana benthamiana. Plant cell tissue organ cult 100:139–148
Tian LM, Huang CL, Zhang XH, Zhang LS, Wu ZY (2005) Advances of plant zinc finger proteins involved in abiotic stress. Biotechnol Bull 6:13–16
Chen WQ, Provart NJ, Glazebrook J, Katagiri F, Chang HS, Eulgem T, Mauch F, Luan S, Zou GZ, Whitham SA, Budworth PR, Tao Y, Xie ZY, Chen X, Lam S, Kreps JA, Harper JF, Si-Ammour A, Mauch-Mani B, Heinlein M, Kobayashi K, Hohn T, Dang JL, Wang X, Zhua T (2002) Expression profile matrix of arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14:559–574
Huang J, Wang JF, Zhang HS (2004) Structure and function of plant C2H2 zinc finger protein. Hereditas (Beijing) 26(3):414–418
Miller J, Mclachlan AD, Klug A (1985) Repetitive zinc-binding domains in the protein transcription factor 111A from xenopus oocytes. EMBOJ 4:1609–1614
Nakashima K, Ito Y, Yamaguchi-Shinozaki K (2009) Transcriptional regulatory networks in response to abiotic stresses in arabidopsis and grasses. Plant Physiol 149:88–95
Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low temperature, or high-salt stress. Plant Cell 6:251–264
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Dong XN (1996) SA, JA, ethylene, and disease resistance in plants. Curr Opin Plant Biol 1:316–323
Lin R, Zhao W, Meng X, Wang M, Peng Y (2007) Rice gene OsNAC19 encodes a novel NAC-domain transcription factor and responds to infection by Magnaporthe grisea. Plant Sci 172:120–130. doi:101016/j.plantsci.2006.07.019
Acknowledgment
The project was supported by the National Natural Science Foundation of China (grants No. 30771477 and No. 30571277).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.






11033_2012_1542_MOESM6_ESM.jpg
Supplementary Data Fig. 2 (a) Nucleotide sequence of ZF30912.1 and its amino acid sequence. (b) Comparison of the amino acid sequence of ZF30912.1. The gene sequence of ZF 30912.1 was compared with 12 ZF with known functions; their accession numbers are as follows: Capsicum annuu CAZFP1 (AT196704.1), Glycine max SCOF1 (GMU68763), Petunia ZPT2-1 (X60700), Arabidopsis thaliana AZF1 (NM126145), Brassica rapa BCZFP1 (AAB53261), Petunia × hybrida ZPT3-3 (AB035133), Petunia × hybrida ZPT2-10 (BAA96070), Arabidopsis thaliana AZF3 (NM-123683), Arabidopsis thaliana AZF1 (NM-001125191), Petunia ZPT2-2 (D26038), Oryza sativa Japanica Group OSZFP34 (AAPA2273), Silene latifolia ZPT3-2 (DQ017765), and Carica papaya ZF30912.1 (gb|ABIM01030863.1|). (JPEG 182 kb)

Rights and permissions
About this article
Cite this article
Jiang, L., Pan, Lj. Identification and expression of C2H2 transcription factor genes in Carica papaya under abiotic and biotic stresses. Mol Biol Rep 39, 7105–7115 (2012). https://doi.org/10.1007/s11033-012-1542-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-1542-y