Abstract
The association between single-nucleotide polymorphisms (SNPs) of the CYP1B1 gene and lung cancer risk is still ambiguous. In this meta analysis, we assessed 10 case–control studies included 7,067 cases and 9,374 controls of the association between CYP1B1 SNPs of Leu432Val (rs1056836, 432C>G), Asn453Ser (rs1800440, 453A>G), Ala119Ser (rs1056827, 119G>T), Arg48Gly (rs10012, 48C>G) and the risk of lung cancer. Crude odds ratios (ORs) with 95% confidence intervals (CIs) were used to evaluate the strength of association between the polymorphism and lung cancer risk under codominant model, dominant model and additive model respectively. Although there were limitations, this meta analysis indicated that individuals with 432GG genotype had a 39.7% higher risk of having lung cancer than those with the 432CC genotype, and individuals with the 432G allele had a 26.3% increased risk as well. An increased risk of lung cancer of 2.13 fold was observed in individuals with 119TT genotype. For Arg48Gly, individuals with 48GG genotype had a significantly increased risk of lung cancer compared with individuals with 48CC (OR 3.859; 95% CI 2.536–5.87). Elevated risk of lung cancer were observed in dominant model (OR 2.115; 95% CI 1.653–2.705) as well. The risk of lung cancer was elevated as the frequency of G allele increased in additive model (P = 0.000). For individuals with the polymorphism at codon 453, no evidence of such association was observed. Furthermore, a possible association between the CYP1B1 polymorphism at codon 432 and the lung cancer could be detected in individuals of Caucasian origin, while a negative association was suggested in Asians and African-Americans. An increased lung cancer risk was also found in women with polymorphism at codon 453. These results are supportive for the hypothesis that the CYP1B1 432GG, 119TT and 48GG genotypes are low-penetrance risk factors for developing lung cancer, and further studies are needed to validate these associations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer is the leading cause of death in the world, with an estimated death of 1,178,918 cases and an estimated new case of 1,332,132 worldwide in 2002 [1]. The exact mechanism of lung carcinogenesis is still under investigation. It has been well documented that tobacco smoking as well as environmental tobacco smoke exposure in life-long time is the most important etiological factor for developing lung cancer [2, 3], the risk of lung cancer in smokers is at least 15 times higher than non-smokers [4, 5]. Carcinogens such as polycyclic aromatic hydrocarbons (PAHs), arylamines and quinones are found to be present in the tobacco smoke. However, only 10–15% of lifelong smokers develop lung cancer, this implies that genetic factors are associated with lung cancer susceptibility as well [6, 7].
Carcinogens present in tobacco smoke are mainly metabolized by phase I and phase II xenobiotic metabolizing enzymes. Recently accumulating studies regarding the role of genes encoding these enzymes in the lung cancer susceptibility have been done. Evidences indicate that genetic polymorphisms of these genes can interfere with the balance between metabolic detoxication and activation of some toxicants, and thus they are relevant to individual susceptibility to lung cancer. A highly studied phase I gene is cytochrome P450 1B1 (CYP1B1). It plays a significant role in the oxidation of variety of carcinogens, including PAHs and arylamines in the lung tissue [8]. Studies demonstrated that CYP1B1 is expressed constitutively in extrahepatic tissues such as lung and is in significantly higher levels within the peripheral leukocytes of lung cancer patients versus controls [9, 10].
CYP1B1 gene is located on chr2p22-p21 and there are at least 179 different polymorphism sites in the gene (http://ncbi.nlm.nih.gov/dbSNP). Of the most studied single-nucleotide polymorphisms (SNP), four are reported to result in amino acid substitutions, they are rs10012 (Arg48Gly), rs1056827 (Ala119Ser), rs1056836 (Leu432Val) and rs1800440 (Asn453Ser). To date, no prospective studies have been done to evaluate the relationship between the CYP1B1 polymorphisms and the risk of lung cancer. Some case–control studies have been conducted. Schoket et al. showed that the CYP1B1 Leu432Val variant was associated with higher levels of DNA adducts within white blood cells [11], and another population-based study reported that non-smoking Caucasians with CYP1B1 Leu432Val polymorphism had increased lung cancer risk [12]. However, the results of these studies are confusing rather than conclusive. In order to get a decisive conclusion, we performed this meta-analysis and evaluate the relationship between the polymorphism of CYP1B1 and the risk of lung cancer.
Methods
Publication search
We conducted a search in the Pubmed (last updated to Feb, 16th, 2010) using the following search terms: (“CYP1B1” or “P4501B1”) and (“genetic variant” or “polymorphism”) and “lung cancer”, trying to cover all papers published. All the searched studies were retrieved, and their references were checked as well for other relevant publications. Review articles were also searched to find additional eligible studies. Only studies published in English with full text available were included. For overlapping studies, only the first published one was selected; for republished studies, only the one with the largest sample numbers was included.
Study identification criteria
The following criteria were followed for study identification: (1) describing the diagnoses of lung cancer; (2) evaluating the association between CYP1B1 genotype and the risk of lung cancer in either a case–control study or a cohort study; (3) supplying the specific number of homozygous and heterozygous carriers in cases and controls, odds ratios (ORs), 95% confidence intervals (CIs) or the information that can lead to the results; (4) fulfilling the Hardy–Weinberg equilibrium in the controls (P > 0.01 was eligible); (5) statistically acceptable methods were mentioned for data collection and analysis; (6) only studies published in English.
Data extraction
Following data were collected if available: first author’s surname, publication year, ethnicity (categorized as Caucasian, Asian, African American), demographics, numbers of cases and controls of different genotypes respectively. We did not define any minimum number of patients to include a study in this meta-analysis. Information was carefully extracted from all eligible publications independently by two of the authors (Xu Wenhuan and Zhou Yunhai).
Statistical methods
Crude ORs of lung cancer associated with CYP1B1 polymorphism were evaluated for every study. We first estimated the risk between rare homozygous and common homozygous carriers. Then we calculated the risk using the dominant model (rare allele carriers vs. common homozygous carriers) and additive model. Heterogeneity assumption was checked by the Q-test and heterogeneity was considered significant for P < 0.10. We combined the data using a fixed-effects model (the Mantel–Haenszel method) or a random-effects model (the DerSimonian and Laird method) according to the heterogeneity. In the absence of heterogeneity, these two methods yield identical results. Random effects are more appropriate when heterogeneity is present. The potential publication bias was examined visually in a funnel plot, and the degree of asymmetry was tested using Egger’s test (P < 0.05 considered representative of statistical significance). Finally we also performed sensitivity analysis by omitting each study to find potential outliers. All the calculations were performed using Stata/SE version 10.0 (Stata Corporation, College Station, TX, USA). All P values are two-sided.
Results
Selected studies
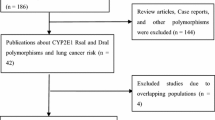
The details of all the studies included were listed in Table 1. We assessed the association between the CYP1B1 polymorphism and the risk of lung cancer via searching the Pubmed. A total of 12 case–control studies [10, 12–22] published as original papers were initially included. Among these reports, six SNPs were analyzed, they were rs10012 (Arg48Gly, 48C>G), rs1056827 (Ala119Ser, 119G>T), rs1056836 (Leu432Val, 432C>G), rs1800440 (Asn453Ser, 453A>G), rs2567206 and rs10916. Since only one study [21] concentrated on rs2567206 and rs10916, we did not perform further analysis about these two polymorphisms. The study from Wu et al. [10] failed to provide details of rare homozygous and heterozygous and therefore was excluded. Cote et al. had published two studies [14, 18] based on subjects enrolled into the same program, therefore we only included the one with the larger number [14]. So a total of 10 publications [12–17, 19–22] including 7,067 cases and 9,374 controls entered the following meta-analysis. Table 2 listed the frequency distribution of the CYP1B1 SNPs Leu432Val, Asn453Ser, Ala119Ser and Arg48Gly. Of those, eight studies [12, 14, 15, 17, 19–22] containing 2,499 cases and 3,199 controls were with regard to the association between the polymorphism of Leu432Val and lung cancer. Four studies [13, 15, 16, 21] with 2,915 cases and 3,791 controls focused on Asn453Ser. With regard to Ala119Ser, three studies [15, 20, 21] with 1,126 cases and 1,816 controls were included. For Arg48Gly, two studies [15, 16] with 527 cases and 568 controls were included. All studies indicated that the distribution of genotypes in controls were consistent with Hardy–Weinberg equilibrium at a significant level of 0.01.
Combined analysis
Table 3 showed the main results on lung cancer risk associated with the CYP1B1 polymorphism of Leu432Val, Asn453Ser, Ala119Ser and Arg48Gly. For Leu432Val, individuals with rare homozygous had a significantly increased risk of lung cancer compared with individuals with common homozygous (OR 1.397; 95% CI 1.172–1.666, P = 0.261 for heterogeneity). The ORs on lung cancer associated with the rare homozygous carriers compared with the common homozygous carriers in individual studies are indicated in Fig. 1. In addition, elevated risk of lung cancer were observed in dominant model (OR 1.263; 95% CI 1.027–1.552, P = 0.011 for heterogeneity). And the risk of lung cancer was elevated as the frequency of G allele increased in additive model (P = 0.000). In terms of SNP Asn453Ser with 2,915 cases and 3,791 controls, no evidence of significant association were observed with the risk of lung cancer in either codominant or dominant model. Elevated risk of lung cancer was found as the frequency of G allele increased in additive model (P = 0.000). With regard to the CYP1B1 SNP Ala119Ser, the eligible studies included 1,126 cases and 1,816 controls. An significant increased risk of lung cancer was found in rare homozygous carriers versus common homozygous carriers (OR 2.130; 95% CI 1.032–4.398, P = 0.023 for heterogeneity) (Fig. 2), while no such association was observed in dominant model (OR 1.414; 95% CI 0.857–2.332, P = 0.000 for heterogeneity). For Arg48Gly, individuals with rare homozygous had a significantly increased risk of lung cancer compared with individuals with common homozygous (OR 3.859; 95% CI 2.536–5.872, P = 0.456 for heterogeneity). Elevated risk of lung cancer were observed in dominant model (OR 2.115; 95% CI 1.653–2.705, P = 0.579 for heterogeneity). All these results were based on pooling data from all the studies regardless of gender, smoking status and ethnicity.
We also stratified the most studied SNP Leu432Val by descent and details were listed in Table 4. Significantly increased risks of lung cancer were found among Caucasian (1,385 cases and 2,084 controls) rare homozygote carriers (OR 1.570; 95% CI 1.049–2.350; P = 0.017 for heterogeneity), while no such association was observed in dominant model (OR 1.400, 95% CI 0.947–2.068; P = 0.002 for heterogeneity). The risk of lung cancer was elevated as the frequency of G allele increased in additive model (P = 0.000). For the Asians with 970 cases and 964 controls, no evidence of significant association were observed with the risk of lung cancer in either codominant or dominant model. There were only two studies dealt with African-Americans and only a total of 144 cases and 151 controls were included. Rare allele carriers seemed not to be associated with lung cancer risk in any model (codominant, dominant, additive model). In addition, we analyzed the influence of gender on the overall OR. In the subgroup of women, the risk of having lung cancer was 37.8% higher in the rare homozygous carriers than the common homozygous carriers (95% CI 1.053–1.803, P = 0.426 for heterogeneity), and the same tendency was observed in dominant model (OR 1.469,95% CI 1.203–1.794, P = 0.565 for heterogeneity) and increased risk of lung cancer was found as the frequency of G allele increased in additive model (P = 0.000).
Sensitivity analysis and publication bias
In addition, we analyzed the influence of individual study on the overall OR of the CYP1B1 SNP Leu432Val (Fig. 3a). We calculated the pooled OR by omitting one single study each time, and no materially huge difference was made. This indicated that there was no individual study affecting the overall OR dominantly. Moreover, we evaluated the publication bias of literatures with Begg’s funnel plot (Fig. 3b). The shape of the funnel plot seems approximately symmetrical, suggesting that publication bias may be neglected. Furthermore, we used the Egger’s test to provide statistical evidence of the symmetry. The Egger’s test is based on a linear regression of the standard normal deviate against its precision, and the intercept value of linear regression measures the symmetry of the funnel plot—the more it deviated from zero, the more pronounced the asymmetry. For the CYP1B1 Leu432Val polymorphism, the egger’s test indicated P = 0.688. This provided evidence that publication bias had no significant effect on the results of our meta-analysis.
Outlier analysis and publication bias analysis for the CYP1B1 Leu432Val polymorphism. a Shows the influence of individual studies on the summary OR. The vertical axis indicates the overall OR and the two vertical axes indicate its 95% CI. Every hollow round indicates the pooled OR when the left study is omitted in this meta-analysis. The two ends of every broken line represent the respective 95% CI. b Shows the Begg’s funnel plot of studies included in the meta-analysis. The vertical axis represents log [OR] and the horizontal axis means the standard error of log [OR]. Horizontal line and sloping lines in funnel plot represent fixed-effects summary OR and expected 95% CI for a given standard error, respectively. Area of each circle represents contribution of the study to the pooled OR the Begg’s funnel plot of studies concerning Leu432Val included in the meta-analysis
Discussion
Lung cancer is the leading frequent and lethal malignant tumor worldwide. For most sporadic lung cancers, a substantial component of risk may be determined by the interaction of multiple low-penetrance susceptibility genes and the exposure to tobacco smoke. It has been accepted that polymorphism of genes involved in the metabolism of tobacco carcinogen are candidate loci for lung cancer susceptibility. Accumulating studies regarding gene variants in the carcinogen metabolism pathway have been performed.
CYP1B1 has been widely studied as a gene encoding phase I metabolizing enzyme. However, evidences of the association between the polymorphism of CYP1B1 and the risk of lung cancer are rather confusing than conclusive. In this meta-analysis, we included 10 publications with a total of 7,067 cases and 9,374 controls. Our result indicated that individuals with the CYP1B1 polymorphisms may contribute to the risk of lung cancer. For SNP Leu432Val, individuals with rare homozygous had a 39.7% higher risk of having lung cancer than those with common homozygous, and increased risk of lung cancer was found in dominant and additive model as well. In addition, for Ala119Ser, a 2.13 fold increased risk of lung cancer was found comparing the rare homozygous carriers with the common homozygous carriers, while no such association was found in dominant model. The risk of lung cancer was elevated as the frequency of T allele increased in the additive model (P = 0.000). For Arg48Gly, an increased risk of lung cancer was found in codominant, dominant and additive model. In addition for SNP Asn453Ser, no evidence of such association was observed in codominant and dominant models.
The exact reason for the correlation between SNPs of CYP1B1 and the risk of lung cancer needs further exploring. Published studies on the structure and functions of the CYP1B1 gene together with their genetic variants may help us understand the potential roles of these polymorphisms. CYP1B1 gene locates on chromosome 2p22-p21 and takes part in the oxidation of procarcinogens, such as PAHs, arylamines [23, 24]. The gene has three exons and two introns [25]. The entire coding sequence is contained in exons 2 and 3, exon 3 has a domain for heme-binding. It is reasonable to conceive that CYP1B1 may affect the metabolism of environmental carcinogens and alter susceptibility to lung cancer. The SNP Ala119Ser is located on exon 2, the putative substrate recognition site 1 [26], and this polymorphism might affect the catalytic activity mediated by CYP1B1 in different individuals. Watanabe et al. reported that the SNP Ala119Ser was associated with the incidence of squamous cell carcinoma of the lung [20]. Jaworowska reported that this polymorphism significantly increase the risk of breast as well as laryngeal cancers in Polish individuals [27]. For the polymorphism Leu432Val at codon 432 of exon 3, it was reported that the 432G allele increased the mutation of p53 [28]. Previous reports demonstrated increased risk of malignant tumors correlated with this polymorphism [29–31]. Consistent with earlier reports on breast cancer [32, 33], individuals with CYP1B1 polymorphism Asn453Ser was not found with increased lung cancer risk in our study.
CYP1B1, in addition to activating procarcinogens in the metabolism of tobacco smoke, is also thought to take part in the hydroxylation of 17β-estradiol. Furthermore, the Leu-Val transition at codon 432 of CYP1B1, which is located in the heme-binding domain of exon 3, causes the enzyme display a higher 4-hydroxylation of estradiol. This leads to the formation of carcinogenic estradiol-3,4-semiquinones and quinones. It is possible that the CYP1B1 is involved in lung carcinogenesis through the estrogenic activity of the 4-OH-E2 [8, 34, 35]. There is possibility that CYP1B1 polymorphism may be associated with lung cancer in woman, since this enzyme is responsible for the oxidizing of procarcinogens in tobacco smoke as well as the formation of carcinogenic metabolic product of estradiol. In our meta-analysis, we found an elevated risk of lung cancer in women carrying the 432 rare allele (OR 1.469; 95% CI 1.203–1.794, P = 0.565 for heterogeneity) as well as the rare homozygous (OR 1.378; 95% CI 1.053–1.803, P = 0.426 for heterogeneity) compared with women carrying common homozygous. Due to the limited number of cases and controls and the lack of detailed information of individuals, we cannot make further analysis according to the menopausal status of women.
For SNP Leu432Val (432C>G), we stratified the result with descent as well. Results indicated that Caucasians with CYP1B1 432GG genotype had significantly increased risk of lung cancer compared with individuals with the CC genotype (OR 1.570, 95% CI 1.049–2.350, P = 0.017 for heterogeneity), while no association was observed in a dominant model (OR 1.400, 95% CI 0.947–2.068, P = 0.002 for heterogeneity). The risk of lung cancer was elevated as the frequency of G allele increased in additive model (P = 0.000). However, the same risk was seen in neither Asians nor African-Americans, indicating that polymorphism of CYP1B1 may be important for individuals with specific ethnicity. Genetic variances in different populations may partially explain this phenomenon. The frequency of 432G allele was varied from 0.378 to 0.558 in Caucasians [12, 14, 19, 21], and it was only 0.087–0.23 in Asians [15, 17, 20, 36]. It was quite surprising that in African-Americans, with the frequency of 432G allele from 0.75 to 0.817 [12, 14, 31], no significant effect was observed with lung cancer risk in codominant, dominant or additive model. This may be due to the limited number of study subjects in these two studies [12, 14].
There are some limitations of this meta analysis. The overall result was based on the crude ORs from pooling of individual unadjusted studies. A more specific result shall be concluded if adjusted by potential suspected factors, such as age and gender. Second, the available data on the polymorphisms were relatively limited, with approximately 2,500 cases and 3,200 controls for Leu432Val, 2,915 cases and 3,791 controls focused on Asn453Ser, 1,126 cases and 1,816 controls for Ala119Ser as well as 527 cases and 568 controls focused on Arg48Gly. This made it quite difficult to perform subgroup analysis. The majority of the subjects presented in our study were cases with lung adenocarcinoma, so the result we concluded may not be representative of all NSCLC’s histological types. There were differences in the original study designs. Most of the studies included were case–control studies while the study by Sorensen et al. [19] was a population based case-cohort one. However, the test of sensitivity did not show materially huge difference while omitting this study. Finally, we did not have detailed individual information on these four SNPs, and this made it impossible to make detailed analyses of linkage disequilibrium and the combined effect of various genetic polymorphisms and lung cancer susceptibility. Furthermore, it is well known that combinations of certain genotypes may be more prominent risk factors compared with single genotype, and large studies including multiple important SNPs of the genes related with the phase I and phase II xenobiotic metabolizing enzymes.
Despite the limitations of this meta-analysis, the overall result of our study suggested that the 432GG genotype, 432G allele, 119TT genotype, 48GG genotype and 48G allele were low-penetrance risk factors for developing lung cancer, while neither T allele at codon 453 nor T allele at codon 119 might alter the individual susceptibility of lung cancer. Furthermore, a possible association between the CYP1B1 Leu432Val polymorphism and the lung cancer could be detected in individuals of Caucasian origin, while a negative association was suggested in Asians and African-Americans. In addition an increased risk of lung cancer was seen in women. However, there is a need for case–control studies and cohort studies to validate the effects of polymorphism at CYP1B1 codon 432, 453, 119 and 48 on metabolizing tobacco smoke carcinogenes in different ethnic groups, different gender and in premenopausal together with postmenopausal women.
References
Parkin DM, Bary F, Ferlay J, Pisani P (2005) Global cancer statistics, 2002. CA Cancer J Clin 55:74–108
Clemens MR (1991) Free radicals in chemical carcinogenesis. Klin Wochenschr 69:1123–1134
Sulhattin A, Sule K, Malik EY, Ozturk O, Ibrahim A (2011) The association between methylene-tetrahydrofolate reductase gene polymorphism and lung cancer risk. Mol Biol Rep 38:991–996
Doll R, Peto R, Wheatley K, Grey R, Sutherland I (1994) Mortality in relation to smoking: 40 years’ observations on male British doctors. Br Med J 309:901–911
Doll R, Peto R (1981) The causes of cancer: quantitative estimates of avoidable risks of cancer in the United States today. J Natl Cancer Inst 66:1191–1208
Hecht SS (1999) Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst 91:1194–1210
Strange RC, Fryer AA (1999) The glutathione S-transferases: influence of polymorphism on cancer susceptibility. IARC Sci Publ 148:231–249
Shimada T, Watanabe J, Kawajiri K, Sutter TR, Guengerich FP, Gillam EM, Inoue K (1999) Catalytic properties of polymorphic human cytochrome P450 1B1 variants. Carcinogenesis 20:1607–1613
Spivack SD, Hurteau GJ, Reilly AA, Aldous KM, Ding X, Kaminsky LS (2001) CYP1B1 expression in human lung. Drug Metab Dispos 29:916–922
Wu MF, Wu WJ, Chang GC, Chen CY, Hu SW, Tsai WT, Lee H, Lin P (2004) Increased expression of cytochrome P4501B1 in peripheral leukocytes from lung cancer patients. Toxicol Lett 150:211–219
Schoket B, Papp G, Levay K, Mrackova G, Kadlubar FF, Vincze I (2001) Impact of metabolic genotypes on levels of biomarkers of genotoxic exposure. Mutat Res 482:57–69
Wenzlaff AS, Cote ML, Bock CH, Land SJ, Santer SK, Schwartz DR, Schwartz AG (2005) CYP1A1 and CYP1B1 polymorphisms and risk of lung cancer among never smokers: a population-based study. Carcinogenesis 26:2207–2212
Rotunno M, Yu K, Lubin JH, Consonni D, Pesatori AC, Goldstein AM, Goldin LR, Wacholder S, Welch R (2009) Phase I metabolic genes and risk of lung cancer: multiple polymorphisms and mRNA expression. PLoS ONE 4:e5652
Cote ML, Yoo W, Wenzlaff AS, Prysak GM, Santer SK, Claeys GB, Van Dyke AL, Land SJ, Schwartz AG (2009) Tobacco and estrogen metabolic polymorphisms and risk of non-small cell lung cancer in women. Carcinogenesis 30:626–635
Shah PP, Singh AP, Singh M, Mathur N, Mishra BN, Pant MC, Parmar D (2008) Association of functionally important polymorphisms in cytochrome P4501B1 with lung cancer. Mutat Res 643:4–10
Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, Stangeland LB, Canzian F, Haugen A (2008) A comprehensive analysis of phase I and phase II metabolism gene polymorphisms and risk of non-small cell lung cancer in smokers. Carcinogenesis 29:1164–1169
Yoon KA, Kim JH, Gil HJ, Hwang H, Hwangbo B, Lee JS (2008) CYP1B1, CYP1A1, MPO, and GSTP1 polymorphisms and lung cancer risk in never-smoking Korean women. Lung Cancer 60:40–46
Cote ML, Wenzlaff AS, Bock CH, Land SJ, Santer SK, Schwartz DR, Schwartz AG (2007) Combinations of cytochrome P-450 genotypes and risk of early-onset lung cancer in Caucasians and African Americans: a population-based study. Lung Cancer 55:255–262
Sørensen M, Autrup H, Tjønneland A, Overvad K, Raaschou-Nielsen O (2005) Genetic polymorphisms in CYP1B1, GSTA1, NQO1 and NAT2 and the risk of lung cancer. Cancer Lett 221:185–190
Watanabe J, Shimada T, Gillam EM, Ikuta T, Suemasu K, Higashi Y, Gotoh O, Kawajiri K (2000) Association of CYP1B1 genetic polymorphism with incidence to breast and lung cancer. Pharmacogenetics 10:25–33
Timofeeva MN, Kropp S, Sauter W, Beckmann L, Rosenberger A, Illig T, Jager B, Mittelstrass K, Dienemann H, Bartsch H, Bickeboller H, Chang-Claude JC, Risch A, Wichmann HE, LUCY-Consortium (2009) CYP450 polymorphisms as risk factors for early-onset lung cancer: gender-specific differences. Carcinogenesis 30:1161–1169
Liang G, Pu Y, Yin L (2005) Rapid detection of single nucleotide polymorphisms related with lung cancer susceptibility of Chinese population. Cancer Lett 233:265–274
Shimada T, Oda Y, Gillam EM, Guengerich FP, Inoue K (2001) Metabolic activation of polycyclic aromatic hydrocarbons and other procarcinogens by cytochromes P450 1A1 and P450 1B1 allelic variants and other human cytochromes P450 in Salmonella typhimurium NM2009. Drug Metab Dispos 29:1176–1182
Shimada T, Fujii-Kuriyama Y (2004) Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci 95:1–6
Stoilov I, Akarsu AN, Sarfarazi M (1997) Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Hum Mol Genet 6:641–647
Gotoh O (1992) Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem 267:83–90
Jaworowska E, Masojc B, Tarnowska C, Brzosko M, Flicinski J, Serrano-Fernandez P, Matyjasik J, Amernik K, Scott RJ, Lubinski J (2006) Association between early-onset breast and laryngeal cancers. Breast Cancer Res Treat 97:215–219
Ko Y, Abel J, Harth V, Brode P, Antony C, Donat S, Fischer HP, Ortiz-Pallardo ME, Thier R, Sachinidis A, Vetter H, Bolt HM, Herberhold C, Bruning T (2001) Association of CYP1B1 codon 432 mutant allele in head and neck squamous cell cancer is reflected by somatic mutations of p53 in tumor tissue. Cancer Res 61:4398–4404
Sasaki M, Tanaka Y, Okino ST, Nomoto M, Yonezawa S, Nakagawa M, Fujimoto S, Sakuragi N, Dahiya R (2004) Polymorphisms of the CYP1B1 gene as risk factors for human renal cell cancer. Clin Cancer Res 10:2015–2019
Sasaki M, Tanaka Y, Kaneuchi M, Sakuragi N, Dahiya R (2003) CYP1B1 gene polymorphisms have higher risk for endometrial cancer, and positive correlations with estrogen receptor alpha and estrogen receptor beta expressions. Cancer Res 63:3913–3918
Tang YM, Green BL, Chen GF, Thompson PA, Lang NP, Shinde A, Lin DX, Tan W, Lyn-Cook BD, Hammons GJ, Kadlubar FF (2000) Human CYP1B1 Leu432Val gene polymorphism: ethnic distribution in African-Americans, Caucasians and Chinese; oestradiol hydroxylase activity; and distribution in prostate cancer cases and controls. Pharmacogenetics 10:761–766
Rylander-Rudqvist T, Wedren S, Granath F, Humphreys K, Ahiberg S, Weiderpass E, Oscarson M, Ingelman-Sundberg M, Persson I (2003) Cytochrome P4501B1 gene polymorphisms and postmenopausal breast cancer risk. Carcinogenesis 24:1533–1539
Bailey LR, Roodi N, Dupont WD, Parl FF (1998) Association of cytochrome P450 1B1 (CYP1B1) polymorphism with steroid receptor status in breast cancer. Cancer Res 58:5038–5041
Li DN, Seidel A, Paritchard MP, Wolf CR, Friedberg T (2000) Polymorphisms in P450 CYP1B1 affect the conversion of estradiol to the potentially carcinogenic metabolite 4-hydroxyestradiol. Pharmacogenetics 10:343–353
Hanna IH, Dawling S, Roodi N, Guengerich FP, Parl FF (2000) Cytochrome P450 1B1 (CYP1B1) pharmacogenetics: association of polymorphisms with functional differences in estrogen hydroxylation activity. Cancer Res 60:3440–3444
Miyoshi Y, Noguchi S (2003) Polymorphisms of estrogen synthesizing and metabolizing genes and breast cancer risk in Japanese women. Biomed Pharmacother 57:471–481
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xu, W., Zhou, Y., Hang, X. et al. Current evidence on the relationship between CYP1B1 polymorphisms and lung cancer risk: a meta-analysis. Mol Biol Rep 39, 2821–2829 (2012). https://doi.org/10.1007/s11033-011-1041-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-1041-6