Abstract
ZEITLUPE (ZTL) plays an important role in the control of flowering time and photomorpogenesis in Arabidopsis and is highly conserved throughout the plant kingdom. Here, we report the characterization of a soybean ZTL homolog GmZTL3 (Glycine max ZTL 3). The absorption spectrum of the recombinant GmZTL3 proteins indicates that it may be a UV/blue photoreceptor. The GmZTL3 expression is independent of diurnal cycles and varies in different tissues along with developmental stages. Before the unifoliolates open fully, GmZTL3 transcripts concentrate in the roots and hypocotyls, while at flowering GmZTL3 accumulates at higher abundance in stems and petioles. Furthermore, the GmZTL3 mRNA accumulates in all kinds of leaves before flowering and concentrates in maturation seeds. In Arabidopsis, the ectopic expression of GmZTL3 delays flowering, implicating GmZTL3 is an inhibitor of flowering induction. Our data indicate that GmZTL3 probably functions as a photoreceptor and plays a role in multiple developmental processes, including the control of flowering time.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The circadian clock regulates 24-h biological rhythms for adapting to the diurnal cycles and temperature fluctuations, which is highly conserved among organisms [1]. The mechanism of operation consists of a series of processes. First, an input pathway perceives and transfers environmental cues relevant to temperature and light to the circadian clock. Second, the endogenous oscillator integrates the environmental cues and transforms them into internal signals. Finally, the output pathway transmits the internal signal into target cells to regulate the physiological and developmental processes [2]. Many physiological and developmental processes, such as cell elongation, stomata opening, leaf movement and flowering are controlled by the circadian clock [1]. The circadian clock consists of one central feedback loop and two lateral feedback loops [3, 4].
In Arabidopsis, ZEITLUPE (ZTL) encodes a 627 amino acid peptide containing three domains: LOV domain, F-box domain and six kelch repeats domain [6]. The ZTL protein was identified as a member of a new photoreceptor family in Arabidopsis, which plays a role in light input to the circadian clock [7, 12]. The ZTL protein is regulated by circadian rhythms and can be recruited to the SCF (Skp1-Cullin-F-box) proteasome complex, which targets degradation of Timing of CAB Expression 1 (TOC1), a component of central oscillator of clock [5, 8]. ZTL can interact with the photoreceptors Cryptochrome 1 (CRY1) and Phytochrome B (PHYB) in Arabidopsis [9], as well as with GIGANTEA (GI) protein and blue light enhances this interaction through the LOV domain [7]. Additionally, the circadian phase-specific degradation of ZTL is mediated by the proteasome and it may be ubiquitinated itself [10]. Moreover, ZTL can negatively regulate the Pseudo-Response Regulator 5 (PRR5) abundance to regulate the circadian oscillator [11].
Currently, there are no reports on ZTL homologs in soybean. In this study, we isolated one ZTL homolog in soybean (Glycine max) and named it GmZTL3 (Glycine max ZTL3). We purified the recombinate GmZTL3 proteins from yeast, measured the absorption spectra, and analyzed the expression profiles of GmZTL3 transcripts in different photoperiods as well as different organs during the developmental progress. Finally we investigated the subcellular localization of GmZTL3 proteins and GmZTL flowering activity through expressing GmZTL3 constitutively in Arabidopsis. The results suggest that GmZTL3 is one ZTL homolog in soybean and involved in control of the flowering process.
Materials and methods
Plant materials and growth conditions
The soybean cultivar Kennong 18 [Glycine max (Merr.) cv. Kennong 18] was grown in artificial climate chambers under both short day (8 h light/16 h dark) and long day conditions (16 h light/8 h dark) at 28°C under a light frequency of 100 μmol m−2 s−1. Seedlings were harvested before the expansion of the unifoliolate leaves. Tissues were sampled independently at the following developmental stages: unifoliolate and 1st, 2nd, 3rd, and 4th trifoliolate fully expanded, and at flowering. Seeds and pods (excluding seeds) were sampled at 7, 14, 21 days after flowering and also at maturity. For circadian analysis, the fully expanded unifoliolates were sampled under short day and long day conditions for two successive 24 h light/dark cycles at 2 h interval. All samples were immediately frozen in liquid nitrogen and stored at −80°C for later use.
Seeds of Arabidopsis wild type C24 and ztl mutants were grown on MS plates for 3 days, grown under long days at 20–22°C for 10 days, and then transplanted to the soil.
RNA isolation, cDNA synthesis and gene cloning
Total RNA was extracted [13] by TRIzol regent (Invitrogen, USA) and cDNA synthesis was performed according to RevertAid first-strand cDNA synthesis kit manual (Fermentas, Germany). The Glymal15g17480 sequence, including UTR and CDS regions, was cloned using RT-PCR with primers of GmZTL3-U-F and GmZTL3-U-R (Table 1). Three independent clones were sequenced, and the resulting consensus sequence was used as the template to amplify the CDS region with primers of GmZTL3-C-F and GmZTL3-C-R (Table 1). The amplicons were inserted into the pGWC vectors [14] and confirmed by sequencing. The yeast expression vector GmZTL3-pYES-DEST52, the ectopic expression vector GmZTL3-pLeela and the subcellular localization vector pEXSG-GmZTL3-YFP were established separately by LR recombination reaction (Invitrogen, USA).
Bioinformatics analysis
The amino acid sequence alignment was carried out using ClustalW software with default parameters, and a Neighbor-joining tree was built using the MEGA software (version 4.0). The bootstrap method was used for assessing the phylogenetic tree, and the number of bootstrap replicates was set to 1000. The search for GmZTL3 LOV domains and pairwise alignment were performed at NCBI using the BLAST network service. The three-dimensional structure of GmZTL3 LOV domain was built using the Swiss Model (http://www.swissmodel.expasy.org) with the Arabidopsis Phototropin2 LOV domain [15, 16] as template (PDB ID: 2Z6D), and the result was analyzed using PyMOL software.
Expression, purification, and absorption spectroscopy measurement of the GmZTL3 protein
The yeast expression vector GmZTL3-pYES-DEST52 was transformed into Saccharomyces cerevisiae strain INVSc1 according to the S.c. EasyCompTM Kit protocol (Invitrogen, USA). Expression of the recombinant fusion protein GmZTL3, which contained polyhistidine (6xHis) tag at the C-terminus, was induced with galactose under dark conditions for 24 h at 28°C. The cells from culture about 1 l was lysed with the breaking buffer (50 mM sodium phosphate, pH 7.4, 5% Glycerol, 1 mM PMSF), using acid-washed glass beads and a bead beater (BiospecProducts, USA). The purification procedure was accomplished following ProBondTM Purification System manual (Invitrogen, USA)under dim red light. The GmZTL3 protein was monitored by anti-His antibody (Abmart, China) and anti-GmZTL raised by our lab in Western blotting. The purified protein was dialyzed against the elution buffer to remove imidazole, which could interfere the absorption spectra of proteins. The purified protein was quantified using a Bradford quantification kit (Bio-Rad, USA). About 100 μg purified GmZTL3 protein was used to measure the absorption spectrum with Back-Man DU-800 spectrophotometer (BeckMan Coulter, USA) at room temperature (25°C) and analyzed with the supporting software and Origin software (Version 7.0). The parameter was set as follows: scan wavelength, 200–800 nm; scan speed, 1200 nm/min; scan interval, 1 nm.
Analysis of gene expression
For GmZTL3 expression analysis, specific primers (Table 1) were used for the quantification of GmZTL3 and GmACT11 (Table 1) was used as the internal control gene [17]. Quantitative real time RT-PCR (qPCR) reactions were performed with the ABI StepOne Detection System (Applied Biosystems, USA). The qPCR reaction consisted of 4 μl template, 7.5 μl 2× SYBR Premix, 300 nM of each primer, and 0.3 μl ROX (TaKaRa, Japan).
Subcellular localization of GmZTL3 proteins
The vector pEXSG-GmZTL3-YFP driven by the Cauliflower mosaic virus 35S promoter and the vector pENSG-CFP-AHL22 served as a maker of nuclear proteins [18] were biolistically co-transformed into Arabidopsis mesophyll protoplasts and the result was recorded by confocal laser scanning microscope (Leica, USA).
Plant transformation
The GmZTL3-pLeela vector was transformed into Arabidopsis C24 and ztl mutant backgrounds via Agrobacterium-mediated transformation. The transgenic lines were screened using 50 mg l−1 glufosinate ammonium and confirmed by PCR with the primers specific to the pLeela and GmZTL3 (Table 1). Three transgenic lines T1 plants (ztl background), together with ten ztl plants, were grown under LD conditions in growth chamber room. T2 plants from three transgenic lines T1 plants (C24 background), together with ten wild-type C24 plants, were grown under the same conditions.
Results
GmZTL3 is a member of the ZTL family
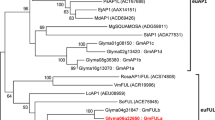
We screened soybean peptide sequences in the Phytozome database (http://www.phytozome.org) using the Arabidopsis ZTL protein sequence as a query. Six peptide sequences were identified which shared three same domains with all AtZTL family: LOV domain, F-box domain and six kelch repeats domain. A further phylogenetic analysis showed four soybean sequences close to AtZTL. Therefore, they were assigned the names GmZTL1, GmZTL2, GmZTL3, and GmZTL4, respectively (Fig. 1). The sequence of GmZTL3, including the UTRs and CDS, was cloned in this study. Alignment of the peptides of GmZTL3 and AtZTL showed a very high similarity and the majority of key residues (including GXNCRFLQ motif) and the spacer of three domains were also conserved (Fig. S1). The CDS of GmZTL3 covered 1,833 bp and encoded a 611-residue polypeptide. The critical amino acids in three domains indicated conserved functions between GmZTL3 and AtZTL.
Phylogenetic tree demonstrating the evolutionary relationships among members of ZTL protein family. The gray shadow indicates the soybean ZTL homologs. Accession numbers: GmFKF1 (Glycine max), Glyma05g34530; GmFKF2 (G. max), Glyma08g05130; GmZTL1 (G. max), Glyma13g00860; GmZTL2 (G. max), Glyma17g06950; GmZTL3 (G. max), Glyma15g17480; GmZTL4 (G. max), Glyma09g06220; OsFKF1 (Oryza sativa), Os011g34460; OsZTL1 (O. sativa), Os06g47890; OsZTL2 (O. sativa), Os02g05700; ZmZTL (Zea mays), GRMZM2G113244; InZTL (Ipomoea nil), ABC25060; McZTL (Mesembryanthemum crystallinum), AAQ73527; AcZTL (Allium cepa), ACT22763; TaZTL (Triticum aestivum), ABR14627; AtZTL, AT5G57360; AtFKF1,AT1G68050; AtLKP2, AT2G18915
Absorption peaks of the GmZTL3 protein appear in UV/blue light region
The three dimensional structure model of the GmZTL3 protein LOV domain shows the analogous hydrophobic structure as the AtPhot2 (Arabidopsis phototropin2) LOV domain, which could bind FMN (flavin mononucleotide) chromophore (Fig. 2b). This result indicated that the GmZTL3 protein may perceive the light. Based on cloning of GmZTL3 into a yeast expression vector with a C-terminal His-tag and galactose induction, the recombinant GmZTL3 protein was purified from yeast (Fig. 2a) and used to analyze the biological activity. As expected, the GmZTL3 protein exhibited a particular spectroscopical characteristic (Fig. 2c). The absorption peaks appeared at 450 nm (blue light region) with a small peak at 340 nm (ultraviolet-A region). These results demonstrated that the GmZTL3 protein possessed properties of a blue-light photoreceptor and may be able to perceive blue light in soybean.
The absorption spectrum of the GmZTL3 protein. a Identification of the purified recombinant GmZTL3 protein monitored by anti-Histidine and anti-GmZTL in Western blotting. b Three dimensional structure model of GmZTL3 LOV domain with the AtPhototropin2 LOV domain as a template (PDB ID: 2Z6D), showing that the three dimensional structure of GmZTL3 LOV domain possesses analogue hydrophobic structure as the Phototropin2 LOV domain. c The absorption spectrum of the GmZTL3 protein
GmZTL3 expression is independent of the diurnal cycle
To study the relationship between the expression of GmZTL3 and light/dark cycles, we tested the mRNA abundance in unifoliolates under long day and short day conditions by quantitative real time RT-PCR (qPCR). The expression level of GmZTL3 kept almost constant excluding a little fluctuation in both LD and SD conditions. Thus the GmZTL3 mRNA shows no obvious circadian rhythm in different photoperiodic conditions in soybean as AtZTL does in Arabidopsis (Fig. 3).
Spatial and temporal expression profiles of GmZTL3
To study the specific expression profiles of GmZTL3 in tissues/organs, we examined the transcripts in most of soybean organs by qPCR. The GmZTL3 transcripts were detectable in different organs and seedlings (Fig. 4). At the unifoliolate stage (vegetative phase), roots accumulated the highest amount of GmZTL3 transcripts. When plants entered the reproductive phase, the stem, the third trifoliolates, and petioles expressed high levels of GmZTL3. As seed development progressed, the GmZTL3 transcript abundance in the pod wall also increased. The data reflected a wide role of GmZTL3 in the broad developmental processes in soybean.
The expression of GmZTL3 in developmental stages of different tissues/organs. Notes for tissue/organs: SL seedlings, R roots, HH hypocotyls, EH epicotyls, C cotyledons, U unifoliolates, SAM shoot apex (including the apical meristem and immature leaves), St stems, L lateral leaves, T1 to T4 the 1st to 4th trifoliolates, Pt petioles, F flower buds, P1 to P3 pod walls at 7, 14, 21 days after flowering, respectively. Notes for developmental stages: Unifoliolates, unifoliolates fully opened; flowering, onset of flowering; seed setting, seed growth. Error bars represents standard deviation of three independent experiments
To elucidate the expression profiles of GmZTL3 during leaf development, we monitored the expression of GmZTL3 in different leaves across different developmental stages. In general, GmZTL3 mRNA accumulated in all kinds of leaves including unifoliolates and different trifoliolates during vegetative growth, but decreased at flowering (Fig. 5a). In seeds, GmZTL3 expression maintained relatively low levels, but increased significantly at maturation (Fig. 5b).
The specific expression of GmZTL3 in leaves (a) and seeds (b) during development. Organs U for unifoliolates; T1 to T4 for the 1st to 4th trifoliolates. Stages at the different leaves (U and T1 to T4) opened fully. Seeds were harvested at 7, 14 and 21 days (7D, 14D and 21D) after flowering or at mature (M). Error bars represent standard deviation of three independent experiments
GmZTL3 proteins localize in both the cytoplasm and nucleus
To visualize the subcellular localization of GmZTL3 proteins, a fusion gene GmZTL3-YFP driven by a 35S promoter was constructed, and then co-transformed with the nuclear marker gene CFP-AHL22 into Arabidopsis leaf protoplasts. The results showed that YFP-associated fluorescence was found in both the cytoplasm and nucleus, while the CFP signal was seen only in nucleus (Fig. 6).
Constitutive expression of GmZTL3 delayed flowering in Arabidopsis
To study the function of GmZTL3 in flowering regulation, we carried out an ectopic expression experiment in both Arabidopsis C24 and ztl mutant backgrounds. The transgenic lines, in both backgrounds, produced more rosette leaves and flowered later in LD conditions (Fig. 7). The transgenic lines T1 plants (ztl background) produced more rosette leaves (Fig. 7a) and flowered later (Fig. 7c) than the ztl plants. The number of rosette leaves produced by the transgenic lines T2 plants in C24 background was also more than their parents (Fig. 7b), and the plants flowered later (Fig. 7d).
Effect of ectopic expression of GmZTL3 on Arabidopsis flowering. a T1 plants carrying 35S::GmZTL3 in ztl mutant, b T2 plants carrying 35S::GmZTL3 in C24 wild type, c the total number of rosette leaves (LN) and days to flowering (DF) corresponding to a, d the total number of rosette leaves (LN) and days to flowering (DF) corresponding to b. Error bars denote the standard deviation, which were from ten ztl plants and three independent T1 plants or ten C24 plants and ten T2 plants derived from three independent T1 plants
Discussion
In Arabidopsis, three different families of blue-light receptors have been characterized: cryptochromes, phototropins, and the ZTL family. LOV domains are highly conserved in all blue light photoreceptors [19]. In our results, the GmZTL3, holding conserved LOV domain, had a similar sequence and structure, and possessed the analogical absorption spectra. Therefore, we speculate that GmZTL3 may act as a photoreceptor to perceive the UV/blue light in soybean.
Similar to ZTL homologs in Arabidopsis, rice and onion [22–24], the GmZTL3 mRNA was constantly expressed independent of photoperiod in soybean. However, the ZTL protein levels oscillate in LD condition in Arabidopsis [10]. The further work to study the relationship between GmZTL3 protein and photoperiod would help us to understand the mechanism of circadian clock in soybean.
In Arabidopsis, ZTL is expressed throughout the plant and in almost all cell types [21]. In this study, we found that GmZTL3 was constitutively expressed throughout the plant. This result is consistent with the argument that GmZTL3 acted as a photoreceptor to participate in the whole plant developmental progress. In previous reports, ZTL plays a role in the process of seedling photomorphogenesis (hypocotyl expansion) in Arabidopsis [25]. The higher expression of GmZTL3 in the root and hypocotyl at the time of the unifoliolates fully opened indicated the GmZTL3 may play a role in morphogenesis in soybean. GmZTL3 may be involved in seed filling due to its higher expression in the mature seed. The expression of GmZTL3 in unifoliolates and different trifoliolates was increased before flowering and then concentrated in the stem and petioles at the time of flowering, inferring that GmZTL3 may function in the control of flowering.
The subcellular localization of transcript regulators contributes to the generation and maintenance of the cellular oscillator in Arabidopsis [20, 21], and the AtZTL protein mobilizes between the nucleus and cytoplasm in circadian-dependent mode [22]. Transient expression analysis revealed the GmZTL3 was expressed in both the cytoplasm and the nucleus, indicating that GmZTL3 played a role as a similar mode as AtZTL does. To examine the effects of clock regulation on GmZTL3 localization would facilitate to elucidate the mechanism of GmZTL function.
In Arabidopsis, ZTL overexpression significantly delays flowering under long-day conditions. Here, the ectopic expression of GmZTL3 in Arabidopsis also delayed flowering independent of plant background, suggesting that GmZTL3 had a similar function in controlling the flowering and that there was no much difference of ZTL function between the short-day plant (soybean) and the long-day one (Arabidopsis). Therefore, the function of ZTL was evolutionally conserved as a flowering inhibitor regardless of the photoperiod response of various plants.
References
Somers DE (1999) The physiology and molecular bases of the plant circadian clock. Plant Physiol 121(1):9–20
Salome PA, McClung CR (2004) The Arabidopsis thaliana clock. J Biol Rhythm 19(5):425–435
Imaizumi T (2010) Arabidopsis circadian clock and photoperiodism: time to think about location. Curr Opin Plant Biol 13(1):83–89
Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Henriques R, Pruneda-Paz JL, Chua NH, Tobin EM, Kay SA, Imaizumi T (2010) F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22(3):606–622
Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293(5531):880–883
Somers DE, Schultz TF, Milnamow M, Kay SA (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101(3):319–329
Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449(7160):356–360
Mas P, Kim WY, Somers DE, Kay SA (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426(6966):567–570
Jarillo JA, Capel J, Tang RH, Yang HQ, Alonso JM, Ecker JR, Cashmore AR (2001) An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature 410(6827):487–490
Kim WY, Geng R, Somers DE (2003) Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc Natl Acad Sci USA 100(8):4933–4938
Kiba T, Henriques R, Sakakibara H, Chua NH (2007) Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19(8):2516–2530
Somers DE, Kim WY, Geng R (2004) The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16(3):769–782
Suzuki Y, Mae T, Makino A (2008) RNA extraction from various recalcitrant plant tissues with a cethyltrimethylammonium bromide-containing buffer followed by an acid guanidium thiocyanate-phenol-chloroform treatment. Biosci Biotechnol Biochem 72(7):1951–1953
Chen QJ, Zhou HM, Chen J, Wang XC (2006) Using a modified TA cloning method to create entry clones. Anal Biochem 358(1):120–125
Lambert LNC, De Bolle X, Depiereux E (2002) ESyPred3D: prediction of proteins 3D strctures. Bioinformatics 18(9):7
Nakasako M, Zikihara K, Matsuoka D, Katsura H, Tokutomi S (2008) Structural basis of the LOV1 dimerization of Arabidopsis phototropins 1 and 2. J Mol Biol 381(3):718–733
Hu R, Fan C, Li H, Zhang Q, Fu YF (2009) Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol Biol 10:93
Xiao C, Chen F, Yu X, Lin C, Fu YF (2009) Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol Biol 71(1–2):39–50
Briggs WR (2007) The LOV domain: a chromophore module servicing multiple photoreceptors. J Biomed Sci 14(4):499–504
Harmer SL, Panda S, Kay SA (2001) Molecular bases of circadian rhythms. Annu Rev Cell Dev Biol 17:215–253
Young MW, Kay SA (2001) Time zones: a comparative genetics of circadian clocks. Nat Rev Genet 2(9):702–715
Kiyosue T, Wada M (2000) LKP1 (LOV kelch protein 1): a factor involved in the regulation of flowering time in Arabidopsis. Plant J 23(6):807–815
Murakami M, Tago Y, Yamashino T, Mizuno T (2007) Comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol 48(1):110–121
Taylor A, Massiah AJ, Thomas B (2010) Conservation of Arabidopsis thaliana photoperiodic flowering time genes in onion (Allium cepa L.). Plant Cell Physiol 51(10):1638–1647
Kim WY, Hicks KA, Somers DE (2005) Independent roles for EARLY FLOWERING 3 and ZEITLUPE in the control of circadian timing, hypocotyl length, and flowering time. Plant Physiol 139(3):1557–1569
Acknowledgments
We thank Drs. George Coupland and Jane Parker kindly provided destination vectors of pLeela, pENSG-GFP, and pEXSP-YFP, and Mr. Feng Zhao raised the GmZTL antibody. This work was supported in part by Transgenic program (Nos 2011ZX08009-001 and 2011ZX08004-005), the National Natural Science Founds (31000681), and the Chinese National Key Basic Research “973” Program (2010CB125906).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Zheng-Gang Xue, Xiao-Mei Zhang and Chen-Fang Lei contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xue, ZG., Zhang, XM., Lei, CF. et al. Molecular cloning and functional analysis of one ZEITLUPE homolog GmZTL3 in soybean. Mol Biol Rep 39, 1411–1418 (2012). https://doi.org/10.1007/s11033-011-0875-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-0875-2