Abstract
The bone morphogenetic protein receptor IB (BMPR-IB) was studied as a candidate gene for the prolificacy of sheep. Nine pairs of primers (P1–P9) were designed to detect single nucleotide polymorphisms (SNPs) of exons 1–4 and 6–10 of the BMPR-IB gene in both high (Small Tail Han and Hu sheep) and low prolificacy breeds (Texel and Chinese Merino sheep) by polymerase chain reaction (PCR)-single strand conformation polymorphism (SSCP). Only the products amplified by primers P2, P5, P6, P7, P8 and P9 displayed polymorphisms. The present study identified 22 SNPs in partial coding regions of ovine BMPR-IB, in which 20 SNPs were reported for the first time. In total of the 22 mutations, 18 DNA variations were originated from the Hu breed, three were found in the Small Tail Han breed (two of them were found in other sheep breeds), three in the Chinese Merino breed, and none in the Texel breed. These results preliminarily demonstrated that BMPR-IB is a major gene affecting the hyperprolificacy in Small Tail Han and Hu sheep, and could be used as a molecular genetic marker for early auxiliary selection for hyperprolificacy in sheep.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ewes from the Booroola strain of Australian Merino sheep are characterized by high ovulation rate and litter size. The Booroola gene (FecB) is an autosomal mutation identified on the basis of segregation studies on litter size [1] and ovulation rate [2]. FecB was the first major gene for prolificacy identified in sheep. The FecB locus is situated in the region of ovine chromosome 6 corresponding to the human chromosome 4q22–23 that contains the bone morphogenetic protein receptor IB (BMPR-IB) gene, which encodes a member of the transforming growth factor β (TGFβ) receptor family [3, 4]. One point mutation at base 746 of the coding region (746A→G) in the highly conserved intracellular kinase signaling domain of the BMPR-IB caused an amino acid change (249Q→R) was associated fully with the hyperprolific phenotype of Booroola ewes [3–5].
The Small Tail Han and Hu sheep are excellent local breeds in China for their significant characteristics of hyperprolificacy and year-round estrus. The mean litter sizes alive of Small Tail Han, Hu, Chinese Merino and Texel sheep have been reported to be 2.61 [6], 2.29 [6], 1.23 [6] and 1.41 [7], respectively. Based on the crucial role of BMPR-IB gene in the regulation of terminal folliculogenesis and the control of ovulation rate, BMPR-IB was considered as a possible candidate gene for prolificacy of sheep. The objectives of the present study were firstly to detect single nucleotide polymorphisms (SNPs) of exons 1–4 and 6–10 of BMPR-IB gene in high (Small Tail Han and Hu sheep) and low prolificacy breeds (Texel and Chinese Merino sheep) by PCR-SSCP, and secondly to investigate the association between BMPR-IB gene and prolificacy in Small Tail Han sheep in which the polymorphisms are segregating.
Materials and methods
Animals
All procedures involving animals were approved by the animal care and use committee at the respective institution where the experiment was conducted. All procedures involving animals were approved and authorized by the Chinese Ministry of Agriculture.
Venous jugular blood samples (10 ml per ewe) were collected from 140 Small Tail Han ewes lambed in 2006, along with data on litter size in the first, second, or third parity (Jiaxiang Sheep Breeding Farm, Shandong Province, China), 40 Chinese Merino ewes (Ziniquan Breeding Sheep Farm, Shihezi City, Xinjiang Uygur Autonomous Region, China), 40 Texel ewes (HITEK Ranch [Beijing] Ltd. Co., Beijing, China) and 38 Hu ewes (Yuhang Hu Sheep Breeding Farm, Hangzhou, China) using acid citrate dextrose as an anticoagulant. Genomic DNA was extracted by phenol–chloroform method, and dissolved in TE buffer (10 mmol/l Tris–HCl [pH 8.0], 1 mmol/l EDTA [pH 8.0]) and kept at −20°C.
The 140 Small Tail Han ewes were selected at random and they were the progeny of five rams. Because the five rams had been sold, their blood was not collected for genotyping. No selection on litter size or other fertility traits was performed in the flock over previous years. Lambing seasons consisted of 3 month groups starting with March through to May as season 1 (spring), June through to August as 2 (summer), September through to November as 3 (autumn) and December through to February as 4 (winter).
Primers and PCR amplification
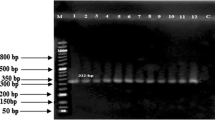
Nine pairs of primers were designed according to mRNA sequence (GenBank accession AF357007) of ovine BMPR-IB. Exons 1–4 and 6–10 of BMPR-IB were amplified. Primer sequence, amplified region, product size and annealing temperature were listed in Table 1.
PCR were carried out in 25 μl volume containing approximately 1.0 μl of 10 μmol/l each primer, 2.5 μl of 10× PCR buffer (50 mmol/l KCl, 10 mmol/l Tris–HCl [pH 8.0], 0.1% Triton X-100), 1.5–1.8 μl of 25 mmol/l MgCl2, 2.5 μl of 2.5 mmol/l each dNTP, 3.0 μl of 50 ng/μl genomic DNA, 1.0 μl of 2.5 U/μl Taq DNA polymerase (Promega, Madison, WI, USA), and the rest is ddH2O. Amplification conditions were as follows: initial denaturation at 94°C for 8 min; followed by 32 cycles of denaturation at 94°C for 30 s, annealing for 30 s (annealing temperature in Table 1), extension at 72°C for 30 s; with a final extension at 72°C for 10 min on Mastercycler® 5333 (Eppendorf AG, Hamburg, Germany).
Single strand conformation polymorphism detection and cloning and sequencing
These steps followed the method of Yan et al. [8].
Statistical analysis
The following fixed effects model was employed for analysis of litter size in Small Tail Han ewes and least squares mean was used for multiple comparisons in litter size among different genotypes.
where y ijklm is the phenotypic value of litter size; μ is the population mean; S i is the fixed effect of the ith sire (i = 1, 2, 3, 4, 5); LS j is the fixed effect of the jth lambing season (j = 1, 2, 3, 4); P k is the fixed effect of the kth parity (k = 1, 2, 3); G l is the fixed effect of the lth genotype (l = 1, 2, 3) and e ijklm is the random residual effect of each observation. Analysis was performed using the general linear model procedure of SAS (ver 8.1) (SAS Institute Inc., Cary, NC, USA). Mean separation procedures were performed using a least significant difference test.
Results
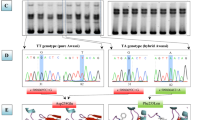
Only the PCR products amplified by primers P2, P5, P6, P7, P8 and P9 displayed polymorphisms. Four genotypes (AA, AC, CC and DD) were detected for primer P2, three (++, B+ and BB) for primer P5, five (GG, GH, HH, KK and LL) for primer P6, three (MM, MN and PP) for primer P7, two (QQ and RR) for primer P8, and two (UU and ZZ) for primer P9.
Polymorphic sequence variations in BMPR-IB in four sheep breeds were showed in Table 2. The 22 SNPs were identified in partial coding regions of ovine BMPR-IB gene in the present study, in which 20 SNPs were reported for the first time. In total of the 22 mutations, 18 base variations were originated from the Hu breed, 3 were found in the Small Tail Han breed (two of them were found in other sheep breeds), 3 in the Chinese Merino breed, and none in the Texel breed.
Except for genotypes MM, MN and PP, amino acid sequences of genotypes AA, ++, GG, QQ and UU were the same as that of ovine BMPR-IB gene published in GenBank (AF357007).
The frequencies of 19 genotypes and 16 alleles in four sheep breeds were presented in Table 3. As for primers P2, P5, P6, P7, P8 and P9, the genotypes and alleles were 4, 3, 5, 3, 2, 2 and 3, 2, 4, 3, 2, 2, respectively. These results indicated that the polymorphisms in coding region of BMPR-IB gene were rather abundant among these sheep breeds.
The test result of difference for BMPR-IB genotype distribution of primer P5 in four sheep breeds was summarized in Table 4. There was highly significant difference (P < 0.001) in the BMPR-IB genotype distribution between high (Small Tail Han and Hu sheep) and low prolificacy breeds (Texel and Chinese Merino sheep). And in either group, no significant difference (P > 0.05) was monitored.
Litter size was significantly influenced by sire, lambing season and parity (P < 0.05, P < 0.05, and P < 0.05, respectively). The least squares means (LSM) and standard errors for litter size of different genotypes of BMPR-IB in Small Tail Han sheep were given in Table 5. For primer P2, the differences of the LSM for litter size between AA, AC and CC were not significant (P > 0.05). For primer P5, the ewes with genotype BB or B+ had 1.51 (P < 0.001) or 1.02 (P < 0.001) lambs more than those with genotype ++; the ewes with genotype BB had 0.49 (P < 0.01) lambs more than those with genotype B+. For primer P6, the differences of the LSM for litter size between GG, GH and HH were not significant (P > 0.05).
Discussion
Polymorphisms of ovine BMPR-IB gene
The FecB mutation (Q249R) is present in Booroola Merino (Australia) [3–5], Garole (India) [9], Javanese (Indonesia) [9], Small Tail Han (China) [8, 10, 11, 14–17], Hu (China) [8, 11–14, 18], and Kendrapada sheep (India) [19]. This study also showed that the FecB mutation is present in Small Tail Han and Hu sheep (China). Therefore, these six ovine breeds may share a common ancestor.
Souza et al. (2001) found two point mutations (A746G and C1113A) by screening 20 Scottish Blackface Merino cross ewes of Booroola genotypes in the kinase domain of BMPR-IB [5]. The present study also identified these two point mutations. A746G mutation was in both Small Tail Han and Hu sheep, not in Texel or Chinese Merino sheep. C1113A mutation was only in Hu sheep, not in other three sheep breeds.
The present study identified 22 SNPs in exons 1–4 and 6–10 of ovine BMPR-IB. Among them, four SNPs (A746G, G922T, T1043C, G192A) led to the change of amino acids (Table 2). According to the known structure of protein kinase genes, the first SNP located between β2 and β3 sheets in nucleotide (ATP)-binding lobe, and the others located in substrate-binding lobe. Specifically, the latter three SNPs situated, respectively, in the subdomain 3, 6, 8 [20] of kinase domain of BMPR-IB.
In two high (Small Tail Han and Hu sheep) and six low fecundity breeds (Dorset, Texel, German Mutton Merino, South African Mutton Merino, Chinese Merino and Corriedale sheep), 17 of 21 SNPs in Inhibin βA gene were from the Hu sheep breed [21]. In the current study, 18 of 22 SNPs were from the Hu sheep breed. The Hu sheep breed is a special local breed in China. Chinese Hu sheep are used mainly for lambskin production and may be found throughout the Taihu Lake area that covers Jiangsu province, Zhejiang province and the vicinity of Shanghai. This sheep breed is famous for its beautiful lambskin, sexual precocity, year-round estrus, and hyperprolificacy [6, 22].This is why so many variations in the Hu sheep breed deserve further study. For primers P2, P6, P8 and P9, no heterozygotes were reported in the Hu sheep breed. Possible reasons include: (1) Heterozygotes exist in the Hu sheep breed, but infertile heterozygotes were probably rapidly removed from the population, or heterozygotes suffered from distinct developmental and reproductive defects, they suffered from the embryonic death, or they were sifted out before arriving at reproductive age. (2) Heterozygotes do not exist in the Hu sheep breed. To verify this hypothesis, a more comprehensive sample should be reported in the Hu sheep.
Effect of BMPR-IB mutation on sheep litter size
The effect of FecB gene is additive for ovulation rate (the number of ova shed at each ovulatory cycle) and partially dominant for litter size. One copy increases ovulation rate by 1.3–1.6 and two copies by 2.7–3.0; litter size is increased by 0.9–1.2 in ewes carrying a single copy and 1.1–1.7 in ewes with two copies [1, 2].
In the BB or B+ ewes, Q249R substitution would impair the inhibitory effect of BMPR-IB on granulosa cell steroidogenesis, leading to their advanced differentiation and an advanced maturation of follicles [3]. This A746G mutation could cause the phenotype characteristic of the Booroola animals which is the precocious development of a large number of small antral follicles resulting in increased ovulation rate [5]. The results in the present study showed that the mean litter size of BB, B+ and ++ genotypes was 2.65, 2.16 and 1.14 in Small Tail Han ewes, respectively. The fact that the FecB mutation is present in prolific Small Tail Han and Hu sheep will promote us to develop breeding strategies to maximize the benefits of increased prolificacy in these breeds and their crosses.
References
Piper LR, Bindon BM (1982) The Booroola Merino and the performance of medium non-Peppin crosses at Armidale. The Booroola Merino. CSIRO, Melbourne, pp 9–20
Davis GH, Montgomery GW, Allison AJ, Kelly RW, Bray AR (1982) Segregation of a major gene influencing fecundity in progeny of Booroola sheep. N Z J Agric Res 25:525–529
Mulsant P, Lecerf F, Fabre S, Schibler L, Monget P, Lanneluc I, Pisselet C, Riquet J, Monniaux D, Callebaut I, Cribiu E, Thimonier J, Teyssier J, Bodin L, Cognie Y, Chitour N, Elsen JM (2001) Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Merino ewes. Proc Natl Acad Sci USA 98:5104–5109
Wilson T, Wu XY, Juengel JL, Ross IK, Lumsden JM, Lord EA, Dodds KG, Walling GA, McEwan JC, O’Connell AR, McNatty KP, Montgomery GW (2001) Highly prolific Booroola sheep have a mutation in the intracellular kinase domain of bone morphogenetic protein IB receptor (ALK-6) that is expressed in both oocytes and granulosa cells. Biol Reprod 64:1225–1235
Souza CJ, MacDougall C, Campbell BK, McNeilly AS, Baird DT (2001) The Booroola (FecB) phenotype is associated with a mutation in the bone morphogenetic receptor type 1B (BMPRIB) gene. J Endocrinol 169:R1–R6
Tu YR (1989) The sheep and goat breeds in China. Shanghai Science and Technology Press, Shanghai
Casas E, Freking BA, Leymaster KA (2004) Evaluation of Dorset, Finnsheep, Romanov, Texel, and Montadale breeds of sheep: II. Reproduction of F1 ewes in fall mating seasons. J Anim Sci 82:1280–1289
Yan YD, Chu MX, Zeng YQ, Fang L, Ye SC, Wang LM, Guo QK, Han DQ, Zhang ZX, Wang XJ, Zhang XZ (2005) Study on bone morphogenetic protein receptor IB as a candidate gene for prolificacy in Small Tailed Han sheep and Hu sheep. J Agric Biotechnol 13:66–71
Davis GH, Galloway SM, Ross IK, Gregan SM, Ward J, Nimbkar BV, Ghalsasi PM, Nimbkar C, Gray GD, Subandriyo Inounu I, Tiesnamurti B, Martyniuk E, Eythorsdottir E, Mulsant P, Lecerf F, Hanrahan JP, Bradford GE, Wilson T (2002) DNA tests in prolific sheep from eight countries provide new evidence on origin of the Booroola (FecB) mutation. Biol Reprod 66:1869–1874
Liu SF, Jiang YL, Du LX (2003) Studies of BMPR-IB and BMP15 as candidate genes for fecundity in Little Tailed Han sheep. Acta Genetica Sinica 30:755–760
Wang GL, Mao X, Davis GH, Zhao ZS, Zhang LJ, Zeng YQ (2003) DNA tests in Hu sheep and Han sheep (small tail) showed the existence of Booroola (FecB) mutation. J Nanjing Agric Univ 26:104–106
Wang QG, Zhong FG, Li H, Wang XH, Liu SR, Chen XJ (2003) The polymorphism of BMPR-IB gene associated with litter size in sheep. Grass-feeding Livestock 2:20–23
Wang QG, Zhong FG, Li H, Wang XH, Liu SR, Chen XJ, Gan SQ (2005) Detection on major gene on litter size in sheep. Hereditas (Beijing) 27:80–84
Davis GH, Balakrishnan L, Ross IK, Wilson T, Galloway SM, Lumsden BM, Hanrahan JP, Mullen M, Mao XZ, Wang GL, Zhao ZS, Zeng YQ, Robinson JJ, Mavrogenis AP, Papachristoforou C, Peter C, Baumung R, Cardyn P, Boujenane I, Cockett NE, Eythorsdottir E, Arranz JJ, Notter DR (2006) Investigation of the Booroola (FecB) and Inverdale (FecXI) mutations in 21 prolific breeds and strains of sheep sampled in 13 countries. Anim Reprod Sci 92:87–96
Jia CL, Li N, Zhao XB, Zhu XP, Jia ZH (2005) Association of single nucleotide polymorphisms in exon 6 region of BMPRIB gene with litter size traits in sheep. Asian-Aust J Anim Sci 18:1375–1378
Yin ZH, Jiang YL, Fan XZ, Wang Y, Tang H, Yue YS (2006) Polymorphism, effect and linkage analysis of BMPR-IB gene in Small Tailed Han sheep. Acta Veterinaria et Zootechnica Sinica 37:510–513
Chu MX, Liu ZH, Jiao CL, He YQ, Fang L, Ye SC, Chen GH, Wang JY (2007) Mutations in BMPR-IB and BMP-15 genes are associated with litter size in Small Tailed Han sheep (Ovis aries). J Anim Sci 85:598–603
Guan F, Liu SR, Shi GQ, Ai JT, Mao DG, Yang LG (2006) Polymorphism of FecB gene in nine sheep breeds or strains and its effects on litter size, lamb growth and development. Acta Genet Sin 33:117–124
Kumar S, Mishra AK, Kolte AP, Dash SK, Karim SA (2008) Screening for Booroola (FecB) and Galway (FecXG) mutations in Indian sheep. Small Rumin Res 80:57–61
Hanks SK, Hunter T (1995) Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 9:576–596
Chu MX, Xiao CT, Fu Y, Fang L, Ye SC (2007) PCR-SSCP polymorphism of inhibin βA gene in some sheep breeds. Asian-Aust J Anim Sci 20:1023–1029
Yue GH (1996) Reproductive characteristics of Chinese Hu sheep. Anim Reprod Sci 44:223–230
Acknowledgments
This work was supported by National Key Technology R and D Program of China (nos. 2008BADB2B01, 2006BAD01A11 and 2006BAD13B08), by the earmarked fund for Modern Agro-industry Technology Research System of China (no. nycytx-39), by National Key Basic Research and Development Program of China (no. 2006CB102105), by Beijing Science and Technology Program of China (nos. Y0705003041131 and BJNY2006-03), by Beijing Natural Science Foundation of China (no. 5042017).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chu, M., Jia, L., Zhang, Y. et al. Polymorphisms of coding region of BMPR-IB gene and their relationship with litter size in sheep. Mol Biol Rep 38, 4071–4076 (2011). https://doi.org/10.1007/s11033-010-0526-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0526-z