Abstract
Genetic variations in the bone morphogenetic protein 15 (BMP15) gene exert a major effect on the ovulation rate in sheep. This study was conducted to investigate the polymorphisms of the BMP15 gene and its association with twining performance in two breeds of Awassi: purebreds and crossbreds. Two loci were genotyped within the exon 2 of the BMP15 gene, B2 and B4 amplicons. PCR–single-strand conformation polymorphism (SSCP) method indicated the presence of two genotypes in B2 amplicons, TT in purebreds and TA in crossbreds. Two novel SNPs were identified in the B2 amplicons, Asp238Glu, which was detected in both studied breeds, and Phe255Leu, which was detected only in crossbreds. Asp238Glu was predicted to have a highly deleterious effect on protein structure and function. Sheep with TA genotype showed 87% of twining ratio compared with sheep with TT genotype that exhibited only 48.3%, signifying an obvious superiority of TA genotype over TT genotype. In conclusions, the non-deleterious Phe255Leu is associated with higher litter size in crossbreds, while the deleterious Asp238Glu is associated with the observed lower litter size on the purebreds. The present study demonstrated the presence of a highly positive effect of Phe255Leu on BMP15 protein, manifesting in a possible improved prolificacy in the ovine population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The mammalian ovulation is a complex trait influenced by several genetic and non-genetic factors. Variation studies in different prolific breeds of sheep showed that one of these genetic factors is the bone morphogenetic protein 15 (BMP15) gene, also known as FecX (Fecundity X gene). The BMP15 gene has a crucial role in regulating the proliferation and differentiation of granulosa cells and plays a key role in the ovulation rate, oocyte quality, and controlling the number of eggs that are ovulated [1]. The BMP15 gene is composed of two exons and is situated on the X chromosome. Genetic variations in the BMP15 gene regulate fertility and affect the reproductive rate in sheep. Several important SNPs have been identified in the BMP15 gene, such as FecXI in Inverdale sheep, FecXH in Hanna sheep [2], FecXB in Irish Belclare sheep, FecXG in Galway sheep [3], FecXL in French Lacaune sheep [4], FecXR in Spanish Rasa Aragonesa sheep [5], FecXGr in French Grivette sheep, FecXO in Polish Olkuska sheep [6], and recently FecXBAR in Tunisian Barbarian sheep [7]. Though these variations differ in type and effect, several natural variations in exon 2 of the ovine BMP15 gene have been shown to cause infertility in homozygous ewes due to defects in the early stages of folliculogenesis [8]. However, the most important variations are likely to be amino acid substitutions that alter the three-dimensional structure of BMP-15 protein, thereby changing its biological function [6]. These variations exhibit the same phenotype: Homozygous carrier ewes are sterile in some prolific breeds of sheep [4], while heterozygous carriers show an increased rate of prolificacy [9]. However, this locus has not been screened in the Awassi breed until now. This breed is a fat-tailed proliferative sheep that characterizes with high capability to withstand harsh environments and is considered as one of the best sheep breeds in the Middle East [10]. This breed is a popular meat-type animal in this area, and genotyping information regarding this breed in relation to the BMP15 gene would be of substantial interest to marker-assisted breeding strategies. Besides, recent revolutions in bio-computational tools that predict the impact of non-synonymous SNPs (nsSNPs) on protein structure and function have not been implemented in this arena to identify potentially hidden issues in this population [11]. Therefore, it is highly mandatory to investigate the possible effect of BMPl5 genetic polymorphisms in the coding portions of the BMP15 protein, as well as to incorporate knowledge of such polymorphisms in the litter size of Awassi sheep. Accordingly, this study was carried out to investigate the presence of polymorphisms in the BMP15 gene in Awassi sheep and its potential association with reproductive traits.
Material and Methods
Sheep Population
This study was conducted according to regulations of the international recommendations for the care and use of animals under the approval of Al-Qasim Green (Agri, No. 015, 3, 12). Careful identification of both included purebred and crossbred Awassi ewes was performed by analyzing the main distinctive morphological characteristics, such as ear length, face morphology, fleece color and distribution, and wool texture [12]. All other sheep having a non-confirmed identity or questioned to have purebred or crossbreeding phenotypes were excluded from the scheduled genotyping experiments. Thus, a total of 184 sexually mature healthy Iraqi Awassi ewes (Ovis aries), consisting of 120 purebreds (Awassi X Awassi) and 64 crossbreds (Awassi X Arrabi), were randomly included in this study. Ewes between 2 and 2.5 years of age were raised in the Barakat Abu al Fadhl Al-Abbas Station (AS) for raising sheep (Al-Khafeel co., Karbala, Iraq). Both maintenance and feeding were similar for all animals and remained under proper animal welfare guidelines for the care and use of livestock animals [13]. Pedigree information of the investigated ewes was updated annually. The recorded relevant ancestor animals were selected in both purebred and crossbred cases. The number of ewes mated per ram varied between 10 and 15. After parturition, ewes were classified into two groups: single lambs and twin lambs.
Genotyping of BMP15 Gene
Venous jugular blood samples (2–3 ml per ewe) were collected from all included sheep. Genomic DNA was extracted from whole blood by a rapid salting-out method [14]. Two specific primer pairs were used to amplify two portions within the exon 2 of the BMP15 gene, B2 and B4 amplicons, of 141 bp and 153 bp, respectively (Fig. 1a). Primer sequences of the B2 amplicon were B2-Hinf1-F-5′-CACTGTCTTCTTGTTACTGTATTTCAATGAGAC-3′ and B2-R-5′-GATGCAATACTGCCTGCTTG-3′, while primer sequences of the B4 amplicon were B4-Dde1-F-GCCTTCCTGTGTCCCTTATAAGTATGTTCCCCTTA and B4-R-TTCTTGGGAAACCTGAGCTAGC-3′ [3]. The PCR was performed using the AccuPower PCR PreMix (Bioneer, South Korea). The PCR program was set as follows: initial denaturation at 94 °C for 4 min, followed by 30 cycles of denaturation (94 °C for 45 s), annealing (63.1 °C for BMP15-B2 and 64.1 °C for BMP15-B4 for 45 s), elongation (72 °C for 45 s), and a final extension (72 °C for 5 min). After performing thermocycling, digestion with restriction enzymes HinfI (5′GANTC) and DdeI (5′CTNAG) was performed for BMP15-B2 and BMP15-B4 amplicons, respectively, to identify previously known variations. Genotyping with the SSCP technique was performed to identify the presence of possible unknown variation(s). The PCR–SSCP was performed according to [11]. Electrophoresis conditions were performed in a vertical slab gel unit according to the manufacturer’s instructions (MiniVS10DSYS, Cleaver Scientific-UK). Electrophoresis reactions were conducted at 8% polyacrylamide at room temperature. Running conditions were set at 200 V, 100 mA for 200 min. DNA fragments in the gels were visualized with PAGE GelRed dye (Biotium, Hayward, USA). Each observed PCR–SSCP pattern was re-amplified and sequenced in forward and reverse directions (Macrogen–Korea). Quality-checked sequences of gene amplicons were aligned by a multiple sequence alignment program according to DNA Star, EditSeq./ClustalW, with sequences published in the GenBank database as a reference to identify polymorphisms. The novelty of SNPs within the BMP15 gene was determined by reviewing the Ensembl genome browser database. Observed variations were deposited in the NCBI nucleotides database (GenBank acc. no. MH938358 and MH938359).
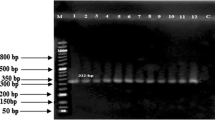
An assessment for the association of BMP15 gene polymorphism with reproductive performance in Awassi sheep. a The genomic positions of two PCR specific primers for B2 and B4 amplicons selected to amplify a portion of exon 2 of the ovine BMP15 gene were described according to GenBank accession no. NC_019484. b PCR–RFLP genotyping of the amplified loci, in which no polymorphism was detected from either B2 or B4 amplicons. c PCR-SSCP genotyping of the same amplified loci, in which the RFLP results were confirmed in B4 amplicons, while B2 amplicon exhibits two genotypes. d DNA sequencing electropherogram of the polymorphic B2 amplicon, in which one SNP, Asp238Glu, was observed in purebred Awassi, and two SNPs, Asp238Glu and Phe255Leu, were observed in the crossbred Awassi. e In silico prediction of both SNPs: Deleterious consequences were observed in Asp238Glu, while neutral consequences were found for Phe255Leu. f The TA genotype of the crossbred Awassi breed exhibited a higher twinning rate than the TT genotype of the purebred Awassi counterpart
Computational Analysis
The amino acid sequence of the ovine BMP15 protein was retrieved from the UniProtKB server (https://www.uniprot.org/uniprot/Q9MZE2). The prediction of the possible deleterious or tolerated effects of observed nsSNPs on BMP15 was performed by SIFT [15], while their potential functional impact was determined using PolyPhen-2 [16]. The previously predicted effects of observed SNPs were validated by PROVEAN [17]. The severity of observed missense SNPs on corresponding amino acid sequences was assessed by the SNAP2 server [18]. Subsequently, the 3D structure of BMP15 was generated by RaptorX server [19] and validated through PhyRe2 mate [20]. The evolutionary conservation status of each detected nsSNP was assessed by the ConSurf tool [21]. The status of each amino acid before and after the missense variation was described within the 3D structure of BMP15 by PyMol-v1, Schrödinger, LLC. The potential roles of observed nsSNPs in posttranslational mechanisms, such as phosphorylation and ubiquitylation, were also predicted by NetPhos3.0 and UbPerd [22, 23]. The putative role of each observed nsSNP in the binding with ligands or receptors was predicted by FTSite and 3DLigandSite tools, respectively [24, 25].
Statistical Analysis
According to the binary nature of litter size, a logistic model with binomial distribution was used in the SAS package (SAS, 2001). The following model was used to investigate the association between litter size and the studied factors in Awassi ewes:
where π is the probability of litter size, α is the scale parameter of the trait, Gi is the fixed effect of the ith genotype (i = TT, TA), Pj is the fixed effect of the jth parity (j = 1, 2, 3, 4), LMk is the fixed effect of the kth lambing month (k = Dec., Nov., and Oct.), and βwt is the covariate factor of the body weight at lambing (64.96 ± 3.66 kg in crossbreds and 55.33 ± 6.16 kg in purebreds). Reproductive traits (fecundity and prolificacy) and twinning ratio were analyzed using the chi-square test, while genotype and allele frequencies and deviation from Hardy–Weinberg equilibrium were evaluated by Popgen32 software, v. 1.31 [26]. The values of polymorphism information content (PIC) were calculated using HET software version 1.8 [27].
Results and Discussion
Results
Genotyping
Initially, the PCR–RFLP method was performed to assess the reported variations in the BMP15 gene. No polymorphisms were identified after digesting B4 (153 bp) amplicons with DdeI restriction endonuclease in both purebreds and crossbreds. Regarding the B2 (141 bp) amplicons, one HinfI heterogeneous pattern of endonuclease digestion was exhibited in all ewes with three discrete electrophoretic bands, including 141, 110, and 31 bp (Fig. 1b), while the potential presence of the unknown variation(s) remains to be confirmed by SSCP protocol. The homogeneity of RFLP digests of B4 amplicons was also confirmed by exhibiting monomorphous SSCP bands. The utilization of SSCP in the B2 amplicons did not confirm the RFLP homogeneity since two electrophoretic bands were detected in purebreds, while the crossbreds showed an extra band in the same specified region in the gel (Fig. 1c). The sequencing reactions confirmed the SSCP electrophoretic difference between the two breeds. Regarding the homozygous SSCP genotypes observed in the purebred Awassi, the sequencing reactions detected one SNP, NC_019484:exon2:c.50980697C > G, or C31G, with a missense effect of Asp238Glu. The same SNP was also observed in the heterozygous SSCP genotypes with an additional SNP, NC_019484:exon2:c.50980646T > A, or T82A, with a missense effect of Phe255Leu. According to T82A SNP, the purebred genotype was assigned TT genotype, while the crossbred was assigned TA genotype (Fig. 1d).
In Silico Prediction
The conducted computational analyses revealed two different changes in the two observed missense SNPs: Asp238Glu and Phe255Leu. The cumulative impact of each SNP was predicted by applying five bio-computational tools: SIFT, PolyPhen-2, PROVEAN, SNAP2, and ConSurf. The amino acid substitution Asp238Glu was found to be deleterious by all in silico tools in terms of structure, function, and evolutionary conservation. However, the Phe255Leu exhibited a completely different computational status in all five computational tools. Phe255Leu was predicted to be entirely non-deleterious on BMP15 structure, function, and evolutionary conservation status (Fig. 1e). Prediction tools were utilized to assess possible participation of both missense SNPs in posttranslational activities of BMP15 protein, such as phosphorylation, ubiquitylation, as well as potential binding with receptors. However, none of these tools suggested any possible role for both detected SNPs in any of these activities.
Reproductive Performance
From 120 ewes of the purebreds, 62 single lambs and 58 twin lambs were produced, while from 64 ewes of the crossbreds only 8 single lambs and 56 twin lamb were produced (Table 1). The total twinning ratio of the purebred Awassi was only 48.3%, while the crossbred ewes exhibited 87.5% of the twinning ratio, which presented a markedly higher value of reproductive performance (Fig. 1f). This observation indicated a significantly higher litter size (P < 0.01) of the sheep with TA genotype than those with TT genotype. Results showed that the BMP15 polymorphism has a significant effect on the litter size (χ2 = 6.576), while no significant association was observed with parity, lambing month, and body weight at lambing month. The observed value of chi-square means indicated that the populations were not in Hardy–Weinberg equilibrium (HWE), which was significant at P < 0.05 (Table 2). Lack of HWE in this population implied that natural or artificial selection favored the mutated allele frequency. According to the classification of PIC (low polymorphism, if PIC value < 0.25; median polymorphism if 0.25 < PIC value < 0.5, and high polymorphism if PIC value > 0.5), the present study showed moderate levels of polymorphism information content within the BMP15 gene.
Discussion
To assess the polymorphism of the BMP15 gene and its possible association with two types of breeds, two coding regions in exon 2 of this gene were considered: B2 amplicons of 141 bp and B4 amplicons of 153 bp. Regarding the absence of any polymorphism in the 153 bp amplicons, our findings are in line with previous studies conducted on several breeds, which indicated no corresponding polymorphism on the DdeI digesting site at 153 bp PCR amplicons [28]. Though RFLP exhibited efficient digestion with HinfI, there were no differences between purebreds and crossbreds, suggesting that all animals were heterozygous in terms of the HinfI recognition site. This finding is not in accordance with the observed differences in the reproductive performance of both Awassi types since the twinning ratio in crossbreds was significantly higher than the purebreds. However, RFLP is still limited in its detection of unknown variations since RFLP is only concerned with the presence or absence of the recognition sequences of the utilized enzymes, while other sequences are not screened in the same amplicons, resulting in loss of variations between the studied ovine species [8]. Instead, the SSCP technique was used due to its powerful sensitivity to detect the potential presence of unknown variation [29]. Therefore, the SSCP technique was employed to resolve this issue by revealing a particular genotype for each type of Awassi breed, in which the purebreds exhibited a different electrophoretic pattern from that found in the crossbreds, suggesting the presence of a particular variation in one of these types that did not exist in another one. Sequencing reactions identified a novel SNP that was positioned in both genotypes. This SNP might be a causal factor for giving all Awassi population one heterozygous RFLP pattern. However, the present study stated that the reason for the presence of two SSCP banding patterns was not attributed to this C31G as it was available in all observed genotypes. Instead, sequencing reactions observed another SNP which was found only in the crossbreds, namely T82A. The latter SNP was not detected by RFLP since it did not have a recognition sequence for the currently utilized HinfI endonuclease. Thus, the reason for the presence of two SSCP genotypes is the presence of T82A SNP which exists only in the crossbreds. A highly deleterious effect for C31G, or Asp238Glu, was identified by all computational tools. Accordingly, this SNP may induce a causal effect in the observed reduced twinning rate in purebreds. Conversely, the cumulative in silico tools indicated the non-deleterious effect of T82A, or Phe255Leu, on the altered protein, suggesting a rather beneficial role for this SNP in giving a remarkably better twinning rate in Awassi sheep. Meanwhile, a beneficial role of Phe255Leu was confirmed by two patterns of experiments, genotyping observations and in silico computations. The genotyping results indicated remarkable superiority of TA genotype for sheep having this SNP, while in silico tools predicted no structural, functional, or evolutionary damaging effects for this alteration. Hence, the approach used to analyze the currently observed SNPs was entirely different from that observed in other ovine BMP15 SNPs [8]. The results from the computational tools on the observed deleterious SNP revealed differences from the previous observation on amino acid 239, i.e., FexG induces a premature stop codon in the place of glutamic acid at amino acid residue 239 of the unprocessed protein, resulting in complete loss of BMP15 function. In contrast, our observed Asp238Glu and Phe255Leu SNPs were found to induce missense effects of respective deleterious and non-deleterious effects in BMP15 protein. Therefore, the resulting BMP15 protein may be normal in its structure, but its function was significantly altered [30]. These BMP15 protein alterations induced by Asp238Glu and Phe255Leu SNPs were reflected by lower or higher twinning rates, respectively.
The present study suggests that the BMP15 heterozygosity described in other breeds has potentially no role in the Awassi breed and that the twinning rate is determined by analyzing the final effects of each observed SNP. This observation was identified from the detection of only two genotypes for the analyzed populations: a pure Awassi with TT (having only C31G SNP) genotype and hybrid Awassi with TA genotype (having both C31G and T82A), while a possible third genotype (AA) was not detected in both populations. The possible explanation behind this point refers to the putative contribution of both genetic patterns in the regulation of litter size in both Awassi populations. This in vitro finding was also supported by several in silico predictions, since these tools have delineated a putative mechanism for these remarkable differences by predicting the effect of both observed SNPs on the final manifestation of the resulting altered protein. Since Asp238Glu SNP is entirely deleterious and Phe255Leu exerted the opposite effect on BMP15, the beneficial Phe255Leu SNP in the crossbred Awassi has potentially alleviated the deleterious Asp238Glu consequences in the purebred Awassi. This possible interaction may be responsible for the higher twinning rate in the crossbreds compared to purebreds.
Conclusion
The present study has identified two novel SNPs: Asp238Glu and Phe255Leu. The former is extremely deleterious and exists in all studied Awassi populations and is responsible for the reduced twinning rate for this population. Phe255Leu is a beneficial SNP observed only in the crossbred Awassi, which may neutralize the deleterious effect of Asp238Glu, leading to improved reproductive regulation and an increased twinning rate.
References
Palai TK, Bisoi PC, Maity A, Behera PC, Sahoo G, Polley S, De S (2013) Prolificacy in Raighar goats is independent of FecB gene. Vet World 6:479–481. https://doi.org/10.5455/vetworld.2013.479-481
Galloway SM, McNatty KP, Cambridge LM, Laitinen MP, Juengel JL, Jokiranta TS, Mc Laren RJ, Luiro K, Dodds KG, Montgomery GW, Beattie AE, Davis GH, Ritvos O (2000) Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nat Genet 25:279–283. https://doi.org/10.1038/77033
Hanrahan JP, Gregan SM, Mulsant P, Mullen M, Davis GH, Powell R, Galloway SM (2004) Mutations in the genes for oocyte-derived growth factors GDF9 and BMP15 are associated with both increased ovulation rate and sterility in Cambridge and Belclare Sheep (Ovis aries). Biol Reprod 70:900–909. https://doi.org/10.1095/biolreprod.103.023093
Bodin L, Di Pasquale E, Fabre S, Bontoux M, Monget P, Persani L, Mulsant P (2007) A novel mutation in the bone morphogenetic protein 15 gene causing defective protein secretion is associated with both increased ovulation rate and sterility in Lacaune Sheep. Endocrinology 148:393–400. https://doi.org/10.1210/en.2006-0764
Monteagudo LV, Ponz R, Tejedor MT, Lavina A, Sierra I (2009) A 17 bp deletion in the bone morphogenetic protein 15 (BMP15) gene is associated to increased prolificacy in the Rasa Araganosa sheep breed. Anim Reprod Sci 110:139–146. https://doi.org/10.1016/j.anireprosci.2008.01.005
Demars J, Fabre S, Sarry J, Rossetti R, Gilbert H, Persani L, Tosser-Klopp G, Mulsant P, Nowak Z, Drobik W, Martyniuk E, Bodin L (2013) Genome-wide association studies identify two novel BMP15 mutations responsible for an atypical hyperprolificacy phenotype in sheep. PLoS Genet 9(4):e1003482. https://doi.org/10.1371/journal.pgen.1003482
Lassoued N, Benkhlil Z, Woloszyn F, Rejeb A, Aouina M, Rekik M, Fabre S, Bedhiaf-Romdhani S (2017) FecXBar a novel BMP15 mutation responsible for prolificacy and female sterility in Tunisian Barbarine Sheep. BMC Genet 18:43. https://doi.org/10.1186/s12863-017-0510-x
Nagdy H, Mahmoud KGM, Kandiel MMM, Helmy NA, Ibrahim SS, Nawito MF, Othman OE (2018) PCR-RFLP of bone morphogenetic protein 15 (BMP15/FecX) gene as a candidate for prolificacy in sheep. IJVSM 6:S68–S72
Davis GH (2005) Major genes affecting ovulation rate in sheep. Genet Sel Evol 37(Suppl. 1):11–23. https://doi.org/10.1051/gse:2004026
Talafha AQ, Ababneh MM (2011) Awassi sheep reproduction and milk production: review. Trop Anim Health Prod 43:1319–1326. https://doi.org/10.1007/s11250-011-9858-5
Al-Thuwaini TM, Al-Shuhaib MBS, Hussein ZM (2020) A novel T177P missense variant in the HSPA8 gene associated with the low tolerance of Awassi sheep to heat stress. Trop Anim Health Prod. https://doi.org/10.1007/s11250-020-02267-w
Al-Shuhaib MBS, Al-Thuwaini TM, Fadhil IA, Aljobouri TRS (2019) GHRL gene-based genotyping of ovine and caprine breeds reveals highly polymorphic intronic sequences in Awassi sheep with several RNA motifs. J Genet Eng Biotechnol 17:3. https://doi.org/10.1186/s43141-019-0004-5
Federation of Animal Science Societies. Guide for the Care and Use of Agricultural Animals in Research and Teaching. Champaign, IL 61822, 3rd edition, 2010.
Al-Shuhaib MBS (2017) A universal, rapid, and inexpensive method for genomic DNA isolation from the whole blood of mammals and birds. J Genet 96:171–176. https://doi.org/10.1007/s12041-017-0750-6
Ng PC, Henikoff S (2006) Predicting the effects of amino acid substitutions on protein function. Annu Rev Genom Hum Genet 7:61–80. https://doi.org/10.1146/annurev.genom.7.080505.115630
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Methods 7(4):248–249. https://doi.org/10.1038/nmeth0410-248
Choi Y, Sims GE, Murphy S, Miller JR, Chan AP (2012) Predicting the functional effect of amino acid substitutions and indels. PLoS One 7:e46688. https://doi.org/10.1371/journal.pone
Calabrese WR, Rudick MM, Simms LJ, Clark LA (2012) Development and validation of Big Four personality scales for the Schedule for Nonadaptive and Adaptive Personality-Second Edition (SNAP-2). Psychol Assess 24:751–763. https://doi.org/10.1037/a0026915
Källberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J (2012) Template-based protein structure modeling using the RaptorX web server. Nat Protoc 7:1511. https://doi.org/10.1038/nprot.2012.085
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:45–858. https://doi.org/10.1038/nprot.2015.053
Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N (2010) ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 38:W529–W533. https://doi.org/10.1093/nar/gkq399
Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S (2004) Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4:1633–1649. https://doi.org/10.1002/pmic.200300771
Radivojac P, Vacic V, Haynes C, Cocklin RR, Mohan A, Heyen JW, Goebl MG, Iakoucheva LM (2010) Identification, analysis and prediction of protein ubiquitination sites. Proteins 78(2):365–380. https://doi.org/10.1002/prot.22555
Wass MN, Kelley LA, Sternberg MJ (2010) 3DLigandSite: predicting ligand-binding sites using similar structures. NAR 38:W469–W473. https://doi.org/10.1093/nar/gkq406
Kozakov D, Grove LE, Hall DR, Bohnuud T, Mottarella SE, Luo L, Xia B, Beglov D, Vajda S (2015) The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat Protoc 10:733–755. https://doi.org/10.1038/nprot.2015.043
Yeh FC, Yang R, Boyle T (1999) POPGENE: version 1.31. Microsoft Window – based freeware for population genetic analysis, University of Alberta. Edmonton, AB, Canada.
Ott J (2001) Program Het version 1.8. Utility programs for analysis of genetic linkage. Rockefeller University. New York, NY, USA
He YQ, Chu MX, Wang JY, Fang L (2006) Polymorphism on BMP-15 as a candidate gene for prolificacy in six goat breeds Chinese. J Anhui Agric Univ 33:61–64. https://doi.org/10.1080/10495390701331114
Hashim HO, Al-Shuhaib MBS (2019) Exploring the potential and limitations of PCR-RFLP and PCR-SSCP for SNP detection: a review. J Appl Biotechnol Rep 6:137–144
Heath DA, Pitman JL, McNatty KP (2017) Molecular forms of ruminant BMP15 and GDF9 and putative interactions with receptors. Reproduction 154:521–534. https://doi.org/10.1530/REP-17-0188
Acknowledgements
The authors are grateful to the staff of Barakat Abu al Fadhl Al-Abbas Station (AS) for raising sheep and to Al-Khafeel co. (Karbala, Iraq) for their facilities that provided the Awassi sheep population. This work did not receive any grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest to publish this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Significance Statement A significant correlation between the BMB15 B2 amplicons and the reproductive performance of Awassi ewes was observed. Thus, these amplicons could be useful markers for assessing ovulation efficiency in Awassi sheep.
Rights and permissions
About this article
Cite this article
Al-Thuwaini, T.M., Aljubouri, T.R.S., Al-Shuhaib, M.B.S. et al. The Effect of Two Novel Amino Acid Substitutions of BMP15 Gene on Ovulation Rate in Awassi Ewes. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 92, 49–55 (2022). https://doi.org/10.1007/s40011-021-01296-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-021-01296-1