Abstract
The erect panicle architecture has contributed to the improvement of yield in japonica rice breeding, and recent molecular analysis has revealed the mechanisms involved in individual plant yield increases. However, the population structure is more important in rice production compared with individual plant yield. Our study compared the population canopy structure of a curved panicle variety Sasanishiki (WT) and an erect panicle mutant derived from CRISPR/Cas9 gene editing at the DENSE AND ERECT PANICLE 1 (DEP1) locus. The results showed that more light could reach to the leaves under the panicle in the CRISPR-dep1 population compared with the WT. The canopy of the CRISPR-dep1 population exhibited higher temperature and lower humidity compared with the WT after heading. A subsequent survey showed that the CO2 concentration in the CRISPR-dep1 population was significantly lower than that in the WT population from full heading to 15 days after heading. Moreover, the increase of biomass in the CRISPR-dep1 population was greater than that in the WT. We noticed that the CRISPR-dep1 mutant could achieve higher yield under low fertilization application compared with the WT under high fertilizer application through increased transplant density. These traits could contribute to an agricultural sustainable development strategy. The quality investigation showed that the dep1 allele increased the yield along with imposing a penalty on grain quality. Our study not only elucidated the mechanism of yield improvement in an erect panicle architecture variety from the perspective of population structure optimization but also provides a theoretical basis for supporting cultivation systems with the erect panicle architecture.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice is one of mankind’s major food staples. Given continuing population growth and increasing competition for arable land between food and energy crops, food security is becoming an ever more serious global problem. Improving crop productivity by selecting the components of grain yield and for optimal plant architecture has been the key focus of national and international rice breeding programs. The past century has witnessed two major breakthroughs in rice breeding. The first was the green revolution of the 1960s in which the development of semi-dwarf lines greatly enhanced the rice yield (Peng et al. 1999b). The second breakthrough was the application of heterosis in hybrid rice, which allowed a continuous increase in rice yield in the 1970s (Virmani et al. 1982; Yuan 1998b). Although these two breakthroughs dramatically improved the rice yield, the imminent food crisis that humanity will face in the next 50 years has forced breeders and scientists to search for the next breakthrough. To achieve this goal, national and international rice breeding programs have emphasized the improvement of crop productivity by selecting superior grain yield components and an ideal plant architecture (Huang et al. 2009; Khush 1999; Peng et al. 2008; Yuan 1998a). Since the 1980s, a number of high-yielding japonica rice strains whose architecture is characterized by dense and erect panicles have been released as commercial varieties.
Recent studies have revealed that the erect panicle architecture is conferred by a heterotrimeric G protein gene DENSE AND ERECT PANICLE 1 (DEP1) that significantly increases the grain yield per plant (Huang et al. 2009; Wang et al. 2009). DEP1 has a modular arrangement with a conventional plant-specific Gγ subunit protein domain at its N-terminus, followed cysteine-rich domain at the C-terminus (Huang et al. 2009). A self-inhibition model was constructed to explain the gain-of-function mechanism that the cysteine-rich domain at the C-terminus has an inhibitory effect on the Gγ protein–like domain at N-terminus (Xu et al. 2016b). Clustered regularly interspaced short palindromic repeats (CRISPR)–associated (Cas) systems have been successfully used as an efficient genome editing tool in a numbers of species (Li et al. 2016; Ma et al. 2015; Shan et al. 2013). The CRISPR/Cas9 gene editing technology was used to verified the function of DEP1 and other important yield-related genes, such as GS3, IPA1, and Gn1a (Li et al. 2016). However, for crops such as rice, maize, and sorghum, the high yield of individual plants does not guarantee a high yield per unit area. The population structure strongly influences the canopy temperature, the circulation of CO2, the humidity, and the amount of sunlight penetration. The population structure of erect panicle varieties has a visual difference compared with normal varieties with curved panicles. The present study surveyed the canopy temperature, the circulation of CO2 and moisture, and the sunlight penetration using the wild-type variety Sasanishiki (WT) and a mutant derived through CRISPR/Cas9 gene editing at the DEP1 locus. These results not only revealed the mechanism of the yield improvement of erect panicle architecture through the perspective of population structure but they also provide a theoretical basis for supporting cultivation system of erect panicle architecture.
Materials and methods
Plant materials
The Japanese japonica variety Sasanishiki (WT) with curved panicles was used in this study as the wild type. Sasanishiki is a typical Japanese variety which was derived from an ancestor variety “Gimbozu,” and widely cultivated in Northeast of China before 1980s. Sasanishiki is a major backbone parent for rice breeding in China, and many currently utilized varieties in Northeast China share the genomic pedigree of Sasanishiki (Fig. S1). Our previous study had generated a DEP1 mutant (CRISPR-dep1) under the genetic background of Sasanishiki using CRISPR/Cas9 gene editing technology (Li et al. 2018). We designed the sgRNA for CRISPR/Cas9 gene editing in the similar region of Shennong265 and Liaojing5 allele (Huang et al. 2009; Wang et al. 2009). Thus, the gene editing plant exhibited similar phenotype of Shenong265 and Liaojing5, as the CRISPR-dep1 allele was also a gain-of-function mutant. We selected the mutant lines that showed erect panicle architecture without Cas9 protein insertion in the genome as mutant plant material (CRISPR-dep1).
Field experiments
All of the materials were grown in the paddy rice field at the Experimental Farm of Shenyang Agricultural University (Shenyang, China; 34° 51 N, 135° 37E) during the summer of 2017 and 2018. The WT and CRISPR-dep1 plants were transplant into field with two levels of nitrogen amount applied and two transplant density treatments across 2 years, using a randomize complete block design with three replicates. Each plot was 40.32 m2. The high-density cultivation was set as 30 cm × 10 cm intervals, and the low-density cultivation used 30 cm × 20 cm intervals. Fertilizers were applied with a basal dressing amount of 60 kg N per hectare, 90 kg P per hectare, and 90 kg K per hectare (low nitrogen fertilization), and 120 kg N per hectare, 90 kg P per hectare, and 90 kg K per hectare (high nitrogen fertilization). Seeds were sown in the seedling nursery on 24 April and transplanted with one seedling per hill on 23 May in both years. The light density, temperature, moisture, and CO2 concentration of the high fertilizers and high-density transplanting treatment between WT and CRISPR-dep1 plots were measured using an iMETOS system (Pessl Instruments GmbH. Werkesweg 107, 8160 Weiz, Austria) following the instruction manual.
Measurements of yield components and quality
To measure the growth rate of the plants, leaf emergence was periodically observed on the main shoot in three plants per plot. The number of tillers of three plants per plot was also counted weekly. At the full heading stage, when 90% of the stems of each plant had emerged from the panicle, the above-ground parts of eight plants were measured from each plot. These samples were then harvested and dried in an oven at 80 °C for 2 days to measure their dry weight. Next, at the mature stage (35 days after the full heading stage), the above-ground parts of 24 plants were harvested from each plot. After counting the panicle number of these plants, the panicles were hand-threshed and put into water. The filled grains, which sank in the water, were separated from unfilled grains, which did not sink in the water. The unfilled grains were further classified into empty (non-fertilized grains) and partially filled grains (poorly developed grains) with the naked eye. Seed sterility was determined by the ratio of non-fertilized grains to total grains. The filled and unfilled grains were then oven dried at 80 °C for 2 days to measure their dry weight. The number of grains per panicle and grain-filling percentage were calculated based on the above data. Three average-sized panicles were taken from each plot to observe the number of primary branches, number of secondary branches, and number of spikelets on each branch. Mature rice grains were milled after harvest, air dried, and stored at room temperature for 3 months. A total of 20 plants from the middle rows were harvested for each line. The brown rice ratio, milled rice ratio, and head rice ratio were calculated after the harvest. The grains were dehulled to produce brown rice using rubber roll sheller (THU testing hunsker, Satake, Hiroshima, Japan), and brown rice ratio was determined. Thereafter, the brown rice was milled with a rice-polishing machine (TM05 test mill, Satake). After milling, head rice and broken rice were separated; finally, the milled rice ratio and head rice ratio were expressed as percentages of the total weight of rough rice. Amylose content and gel consistency were assessed according to The National Standard of the People’s Republic of China (GB/T17891–1999). Extraction and measurement of rice protein composition was performed as described by Tan et al. (1999). All of the samples were analyzed twice.
Statistical analysis
All data were analyzed using excel (2016) software (Microsoft, USA). Data were analyzed following analysis of variance (ANOVA) and means of materials and among treatments within a materials were compared based on the least significant difference (LSD) test at the 0.05 probability level.
Results
Generation of a DEP1 mutant line with CRISPR/Cas9 gene editing technology
In order to compare the differences in population structure of erect panicle and curved panicle architectures caused by the DEP1/dep1 allele, we used the dep1 mutant generated by CRISPR/Cas9 gene editing technology under the genetic background of a famous Japanese japonica variety, Sasanishiki (WT) (Fig. 1). The mutant encoded a similar truncated DEP1 protein as the dep1 allele of SN265 (a widely released japonica commercial variety with erect panicles) (Li et al. 2018). The mutant exhibited erect panicle architecture, shorter panicles, and more grains per panicle compared with the WT. We crossed the mutant to the WT, and the panicle length of the F2 population showed a 1:3 segregation, which confirmed the mutant in dep1 locus corresponding to the erect phenotype (Fig. 1). We scanned the segregating groups and successfully identified a line (CRISPR-dep1) that had no Cas9 protein. We surveyed the plant architecture of the WT and CRISPR-dep1 at full heading stage and full ripeness stage (Fig. 2). At the heading stage, no obvious difference in plant architecture between the WT and the CRISPR-dep1 mutant was observed. Only the angle of third leaf in the WT was significantly larger than that of CRISPR-dep1. At the full ripeness stage, the angle of the panicle in the WT was dramatically increased, whereas the angle of the panicle in CRISPR-dep1 was barely changed. The panicle neck angle of CRISPR-dep1 was approximately 12.3°, whereas in the WT this was around 50°. The panicle length, plant height, culm height, and the length of flag leaf of the WT were significantly longer than those of CRISPR-dep1. There was no significant difference in the angle of the flag leaf between the WT and CRISPR-dep1.
The CRISPR/Cas9 genome edited plant of DEP1. a A CRISPR/Cas9 gene edited plant at the DEP1 locus in pot. Scale bar, 20 cm. b Schematic diagram of the genomic region of DEP1 (Sasanishiki), CRISPR-dep1, and dep1 (Shennong265 and Liaojing5). A self-inhibition model was constructed to explain the gain-of-function mechanism. c Sequence alignment of the sgRNA target region of the WT and CRISPR-dep1 plants. d The panicle length of the F2 population derived from the cross between CRISPR-dep1 and the WT. e The WT and CRISPR-dep1 in the field when the population reach heading. f The WT and CRISPR-dep1 in the field when the population reach mature. g The iMETOS system in the field
The plant architecture of DEP1 and CRISPR-dep1 plants. a The model of angle measurement. P: the angle of the panicle; F: the angle of the flag leaf; 2: the angle of the second leaf; 3: the angle of the third leaf. b The angle measurement of DEP1 and CRISPR-dep1 plants at full heading stage. c The angle measurement of DEP1 and CRISPR-dep1 plants at full ripeness stage. An asterisk denotes significant at the 5% level
The vertical distribution of light in different panicle architecture populations
The photosynthetic ability in the panicle is negligible, but the shadow of the panicle on lower leaves may reduce photosynthetic products. In order to demonstrate the difference in light penetration between the two plant architectures, we surveyed the light density at different canopy heights at noon from full heading stage to full ripeness stage in 2017 and 2018. A similar degree of light penetration was observed between the WT and CRISPR-dep1 at full heading stage in 2018. However, along with panicles beginning to curve in the WT, the light penetration of the WT was less than in CRISPR-dep1 at 15 days after heading. The light density in the CRISPR-dep1 population at 30 cm from the canopy was obviously greater than that of the WT. At full ripeness stage, the light penetration of CRISPR-dep1 enlarged its advantage, and the leaves under the panicle received more light compared with the WT (Fig. 3). A similar trend was observed in 2017 (Fig. S2).
The light penetration in canopy in 2018. a The light penetration from canopy to 45 cm from the canopy in DEP1 population and CRISPR-dep1 population at full heading stage. b The light penetration from canopy to 45 cm from the canopy in DEP1 population and CRISPR-dep1 population at 15 days after heading. c The light penetration from canopy to 45 cm from the canopy in DEP1 population and CRISPR-dep1 population at full ripeness stage
The daily changes in canopy conditions
We subsequently monitored the CO2 concentration, temperature, and humidity of the canopy in two populations (Fig. 4). At full heading stage, the canopy temperatures of the WT and CRISPR-dep1 were similar. From the 15 days after heading to the full ripeness stage, the temperature of CRISPR-dep1 was higher than that of the WT, especially at noon. The canopy humidity of CRISPR-dep1 was lower than that of the WT from heading to maturity. A similar trend was observed in 2017 (Fig. S3). Interestingly, we found that the CO2 concentration of CRISPR-dep1 was significantly lower than that of the WT at full heading stage, and the difference in CO2 between WT and CRISPR-dep1 became lesser from full heading stage to full ripeness stage. At 15 days after heading and ripeness stages, the CRISPR-dep1 population only showed lower CO2 concentration than WT population at midday-noon. These results indicated that the CRISPR-dep1 population might have had a higher photosynthetic rate than that of the WT population at full heading stage. Taken together, the results suggest that the canopy in the CRISPR-dep1 population exhibited lower CO2 concentration, higher temperature, and lower humidity during the filling stage of rice.
The biomass and yield of different plant architecture populations
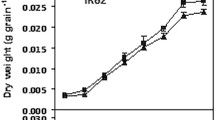
We subsequently investigated the biomass of the WT and CRISPR-dep1 from heading to ripeness using a 14-day interval survey. The results were consistent with the CO2 concentration survey. The CRISPR-dep1 population showed a higher increase rate than that of the WT population. These results implied that the higher photosynthetic rate in the CRISPR-dep1 population from full heading stage to 15 days after heading produced more biomass than that of the WT population. Moreover, we found that the dry weight of panicles of the WT and CRISPR-dep1 were similar at full heading stage. The panicle dry weight of CRISPR-dep1 had a higher increase rate compared with that of the WT. These results indicated that a greater proportion of the biomass increase was accumulated into panicles in the CRISPR-dep1 population compared with the WT. Based on our study and the report from other research team (X et al. 2019), the truncated dep1 allele could significantly reduce the plant height and panicle length, but the effect on grain size was small. The length of grain in CRISPR-dep1 was slightly shorter but wider than that of WT (Fig. 5). We then investigated the yield of the WT and CRISPR-dep1 under different cultivation methods. Two levels of fertilization and two transplanting densities were used in this experiment. The results showed that the high level of fertilization significantly increased the yield per unit of the WT, whereas the high transplant density negatively affected yield of the WT. However, the high level of fertilization also significantly increased the yield per unit of CRISPR-dep1, but the high transplant density positively affected grain yield. The CRISPR-dep1 harvested more grains per unit under the cultivation method of low fertilization and high transplant density than the WT under the cultivation method of high fertilization. The yield components in 2017 also observed similar trend (Table 1). Thus, we could increase the grain yield of CRISPR-dep1 through increasing the transplant density without increasing the amount of fertilizer.
The biomass and yield performance of WT and CRISPR-dep1 plants. a The panicles of the WT and CRISPR-dep1 plants. b The grain numbers per panicle of the WT and CRISPR-dep1 plants. c The grain size of the WT and CRISPR-dep1 plants. d The biomass of the WT and CRISPR-dep1 plants from heading to full ripeness during a 14-day interval survey. e The percentage of panicle to the whole biomass of the WT and CRISPR-dep1 plants from heading to full ripeness during a 14-day interval survey. An asterisk denotes significant at 5% level. Bar = 1 cm
The quality of different plant architecture populations
As the erect panicle varieties were considered to be only of mediocre eating quality, we conducted a quality investigation to elucidate whether dep1 imposed a penalty in regard to quality in rice production (Fig. 6). The results confirmed that the dep1 allele imposed such a penalty in grain quality. The dep1 allele decreased the grain shape (the ratio of grain length to grain width), which subsequently increased the head rice ratio. The dep1 allele also strongly affected chalkiness and related traits. The chalkiness ratio and the chalkiness level were significantly higher in CRISPR-dep1. The CRISPR-dep1 had higher amylose content and protein content compared with those of the WT. We compared the texture properties of cooked rice between the WT and CRISPR-dep1. The results showed that the CRISPR-dep1 had a higher hardness score but a lower stickiness score compared with the WT. CRISPR-dep1 also had a lower taste score than the WT.
Discussion
Breeding to improve plant architecture has tremendously contributed to the increase in rice production. The semi-dwarf varieties greatly enhanced rice yield in the 1960s (Peng et al. 1999a). The ideal plant architecture was based on the foundation of semi-dwarf breeding. Erect panicle architecture has been extensively used in japonica rice breeding (Qian et al. 2016). The northeast area and downstream region of the Yangtze River represent over 70% of the entire japonica rice cultivated area in China. At present, more than 50% of the dep1-carrying cultivars are propagated in the northeast of China. At the downstream region of the Yangtze River, nearly all of the japonica varieties have the erect panicle architecture. Moreover, the cultivation area for erect panicle varieties has annually increased in the japonica zone (Xu et al. 2016a). Although recent molecular studies have demonstrated that the DEP1 locus corresponded to erect panicles and the molecular mechanism of yield increase for individual plants became clear, the contribution of erect panicle architecture to population canopy improvement was unexplored. Since the application of semi-dwarf breeding, the leaf area index (LAI) has significantly increased. However, modern rice breeding is no longer simply the pursuit of high LAI, but rather the enhancement of the population photosynthesis and material production capacity through the improvement of population structure and canopy light reception. As most leaves are below the panicle, the curved panicle at the filling stage may influence population-level photosynthesis. Many lines of evidence indicate that solar radiation has a positive impact on rice growth and development, and solar radiation required by rice crops at different phenological stages differs (Peng et al. 2004; Islam and Morison 1992; Yoshida and Parao 1976). Our study has shown that more light could reach the leaves under the panicle, especially at 15 days after heading in CRISPR-dep1 compared with the WT. Compared with the canopy of the WT, the canopy of CRISPR-dep1 had higher temperatures and lower relative humidity. The higher temperature of the canopy during the last phase of the filling stage might mitigate the effect of low temperature northeast of China, thereby aiding the accumulation of photosynthate. The high humidity of the canopy might lead to increased incidence of diseases and insect pests. The lower humidity of the canopy in CRISPR-dep1 might enhance the resistance to diseases and insect pests. The subsequent survey showed that the CO2 concentration in the CRISPR-dep1 population was significantly lower than that in the WT population from full heading to 15 days after heading. Moreover, the increase in biomass in the CRISPR-dep1 population was higher than in the WT during this period. A higher proportion of biomass was accumulated into panicles in the CRISPR-dep1 population compared with the WT population.
The WT and CRISPR-dep1 populations showed diversity in yield performance under different cultivation methods. As expected, both WT and CRISPR-dep1 exhibited the highest individual plant yield under high fertilizer application and low transplant density cultivation. When we investigated the yield per unit area, we found that the highest yield per unit area for the WT was under high fertilizer application and low transplant density cultivation, whereas the highest yield per unit area for CRISPR-dep1 was achieved under high fertilizer application and high transplant density cultivation. These results indicated that the dense plant architecture of erect panicles improved the population canopy structure, and eventually increased the yield per unit area through increasing the number of plants per unit area. We noticed that the CRISPR-dep1 could achieve higher yield per unit area under low fertilizer application cultivation than the WT under high fertilizer application cultivation through increasing the transplant density. These traits could contribute to a sustainable development strategy. Thus, the erect panicle architecture is another important heritable morphological trait after semi-dwarf breeding and is the ideal plant architecture needed to meet the requirements of super high-yield rice breeding.
CRISPR-associated (Cas) systems have become an efficient genome editing tool in gene function research. As DEP1 plays a crucial role in rice production, the function of DEP1 was verified by several research team using CRISPR/Cas9 technology (Li et al. 2016; Shen et al. 2017; Wang et al. 2017; Li et al. 2019; Sun et al. 2018). The CRISPR/Cas9 gene editing plants could be divided into two types: type I, the sgRNA was designed in a similar region of Shennong265, and Liaogeng5 in exon 5, which only eliminated the Cys-rich region (Li et al. 2016; Li et al. 2019); type II, the sgRNA was designed in the first exon of DEP1 which eliminated both Gγ protein region and Cys-rich region (Sun et al. 2018; Shen et al. 2017). The type I CRISPR/Cas9 gene editing plant showed an erect panicle architecture, an increase in grain number per panicle, and a reduction in plant height, panicle length, and grain length. The type II plants also exhibited an erect panicle architecture, but a reduction in grain number per panicle and setting rate. The phenotype of transgenic plant in present study was similar to type I gene editing plant. Moreover, both type I and type II transgenic plants showed a reduction in grain length which caused a decrease of 1000 grain weight. Our study showed that truncated dep1 (25.43 ± 0.37 g) had a similar 1000 grain weight compare with that of DEP1 (25.58 ± 0.25 g), as the truncated dep1 allele could increase the width of grain (Fig. 5). The present study has demonstrated that an imposed penalty for grain quality was a pleiotropic effect of the dep1 allele. The dep1 allele might increase the amylose and protein contents, leading to higher scores for hardness and lower scores for of stickiness, and eventually cause decreases in the taste score. This result was consistent with erect panicle varieties being of only mediocre eating quality. Thus, high-yield and high-quality erect panicle varieties might be generated through the combination of the dep1 allele and other premium grain quality alleles through rationally designed breeding.
References
Huang X, Qian Q, Zhengbin L, Hongying S, Shuyuan H, Da L, Guangmin X, Chengcai C, Jiayang L, Xiangdong F (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41(4):494–497
Islam MS, Morison JIL (1992) Influence of solar radiation and temperature on irrigated rice grain yield in Bangladesh. Field Crop Res 30(1–2):13–28
Khush GS (1999) Green revolution: preparing for the 21st century. Genome 42(4):646–655
Li M, Xiaoxia L, Zejiao Z, Pingzhi W, Maichun F, Xiaoping P, Qiupeng L, Wanbin L, Guojiang W, Hongqing L (2016) Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front Plant Sci 7
Li X, Tao X, Miao J, Yang Z, Gu M, Liang G, Zhou Y (2019) Evaluation of differential qPE9-1/DEP1 protein domains in rice grain length and weight variation. Rice 12(1):5
Li X, Wu L, Wang J, Sun J, Xia X, Geng X, Wang X, Xu Z, Xu Q (2018) Genome sequencing of rice subspecies and genetic analysis of recombinant lines reveals regional yield- and quality-associated loci. BMC Biol 16(1):102
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8(8):1274–1284
Peng J, Richards Donald E, Hartley Nigel M, Murphy George P, Devos Katrien M, Flintham John E, James B, Fish Leslie J, Worland Anthony J, Fatima P (1999a) Green revolution’genes encode mutant gibberellin response modulators. Nature 400(6741):256–261
Peng S, Cassman Kenneth G, Virmani SS, Sheehy J, Khush GS (1999b) Yield potential trends of tropical rice since the release of IR8 and the challenge of increasing rice yield potential.
Peng S, Jianliang H, Sheehy John E, Laza Rebecca C, Visperas Romeo M, Xuhua Z, Centeno Grace S, Khush Gurdev S, Cassman Kenneth G (2004) Rice yields decline with higher night temperature from global warming. Proc Natl Acad Sci U S A 101(27):9971
Peng S, Khush Gurdev S, Parminder V, Qiyuan T, Yingbin Z (2008) Progress in ideotype breeding to increase rice yield potential. Field Crop Res 108(1):32–38
Qian Q, Longbiao G, Smith Steven M, Jiayang L (2016) Breeding high-yield superior-quality hybrid super-rice by rational design. National Science Review, p nww006
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31(8):686–688
Shen L, Yufeng H, Yaping F, Jian L, Qing L, Xiaozhen J, Gaowei X, Junjie W, Xingchun W, Changjie Y (2017) Rapid generation of genetic diversity by multiplex CRISPR/Cas9 genome editing in rice. Sci China Life Sci 60(5):506–515
Sun S, Lei W, Hailiang M, Lin S, Xianghua L, Jinghua X, Yidan O, Qifa Z (2018) A G-protein pathway determines grain size in rice. Nat Commun 9(1):851
Virmani SS, Aquino RC, Khush GS (1982) Heterosis breeding in rice (Oryza sativa L.). Theor Appl Genet 63(4):373–380
Wang J, Tetsuya N, Shuqian C, Wenfu C, Hiroki S, Takuji T, Yutaka O, Zhenjin X, Takatoshi T (2009) Identification and characterization of the erect-pose panicle gene EP conferring high grain yield in rice (Oryza sativa L.). Theor Appl Genet 119(1):85–91
Wang Y, Lizhao G, Menglong Y, Juan W, Chen J, Min L, Kun Y, Ya Z, Huaibing J, Eric W (2017) Deletion of a target gene in Indica rice via CRISPR/Cas9. Plant Cell Rep 36(8):1–11
Xu H, Zhao M, Zhang Q, Xu Z, Xu Q (2016a) The dense and erect panicle 1 (DEP1) gene offering the potential in the breeding of high-yielding rice. Breed Sci 66(5):659–667
Xu Q, Mingzhu Z, Kun W, Xiangdong F, Qian L (2016b) Emerging insights into heterotrimeric G protein signaling in plants. Journal of Genetics and Genomics
Yoshida S, Parao FT (1976) Climatic influence on yield and yield components of lowland rice in the tropics. Climate & Rice
Yuan LP (1998a) Hybrid rice breeding in China. Advances in Hybrid Rice Technology. Philippines, International Rice Research Institute, pp 27–33
Yuan L (1998b) Hybrid rice breeding for super high yield. 21st century: 10
Funding
The National Natural Science Foundation of China (U1708231) supported this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
Fig. S1 The pedigree of Sasanishiki. Fig. S2 The light penetration of canopy in 2017. (a) The light penetration from canopy to 40 cm from the canopy in DEP1 at full heading stage. (b) The light penetration from canopy to 40 cm from the canopy in CRISPR-dep1 at full heading stage. (c) The light penetration from canopy to 40 cm from the canopy in DEP1 at full ripeness stage. (d) The light penetration from canopy to 40 cm from the canopy in CRISPR-dep1at full ripeness stage. Fig. S3 The temperature and humidity of canopy in 2017. (a) The canopy temperature in the DEP1 and CRISPR-dep1populations at full heading stage. (b) The canopy temperature in the DEP1 and CRISPR-dep1populations at full ripeness stage. (c) The canopy humidity in the DEP1 and CRISPR-dep1populations at full heading stage. (d) The canopy humidity in the DEP1 and CRISPR-dep1populations at full ripeness stage. (PPTX 177 kb)
Rights and permissions
About this article
Cite this article
Fei, C., Yu, J., Xu, Z. et al. Erect panicle architecture contributes to increased rice production through the improvement of canopy structure. Mol Breeding 39, 128 (2019). https://doi.org/10.1007/s11032-019-1037-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-019-1037-9