Abstract
Genetic analysis of a diverse set of 42 traits for flower (5), phenology (9), fruit quality (19), leaf (8) and disease resistance (1) was carried out in two interspecific almond × peach populations, an F2 (T × E) and a BC1 (T1E), from the cross between ‘Texas’ almond and ‘Earlygold’ peach. Traits related to flower, phenology, fruit quality, leaf morphology and resistance to powdery mildew were phenotyped over 3 years in two locations and studied for co-segregation with a large set of SNP and SSR markers. Three maps were used, one for the T × E and two for the T1E (T1E and E) population. Nine major genes were identified and mapped: anther color (Ag/ag and Ag2/ag2), flower color (Fc2/fc2), maturity date (MD/md), almond fruit type (almond vs. peach; Alf/alf), juiciness (Jui/jui), blood flesh (DBF2/dbf2), powdery mildew resistance (Vr3) and flower type (showy/non-showy; Sh/sh). These genes were often located in genome positions different from those for major genes for similar traits mapped before. Two of them explain fundamental aspects that define the fruit of peach with respect to that of almond: Alf and Jui, for its thick and juicy mesocarp, respectively. The genetics of quantitative traits was studied, and 32 QTLs were detected, with consistent behavior over the years. New alleles identified from almond for important traits such as red skin color, blood flesh, fruit weight and powdery mildew resistance may prove useful for the introduction of new variability into the peach gene pool used in commercial breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Peach, Prunus persica L. (Batsch), is an economically important temperate tree fruit and one of the model species for the Rosaceae family (Shulaev et al. 2008). Its genome has been recently sequenced (Verde et al. 2013), and important information on the genetics of major genes and QTLs obtained in the last two decades (Arús et al. 2012). Very early on, geneticists realized that the genetic basis of North American and European commercial cultivars was very narrow (Scorza et al. 1985), later confirmed with molecular markers not only for these materials but also for modern Asian cultivars (Li et al. 2013; Micheletti et al. 2015). The more variable ancient oriental cultivars and landraces could be a source of new alleles for peach breeding programs (Li et al. 2013; Cao et al. 2014). An additional major source of new genetic variability is the close, cross-compatible relatives of peach, such as almond [P. dulcis (Miller) D.A. Webb] and several wild species (P. davidiana Carr., P. cerasifera Ehrh., P. mira Koehne and P. kansuensis Rehder). Interspecific hybrids between peach and these species have been used for rootstock development (Byrne et al. 2012), and for genetic analysis, looking for resistance to pests and diseases such as green peach aphid, plum pox virus, powdery mildew and nematodes (Gradziel 2003; Foulongne et al. 2003; Claverie et al. 2004; Rubio et al. 2010; Sauge et al. 2012). They have also been used to analyze fruit quality (Gradziel 2003; Quilot et al. 2004) and tree architecture (Carrillo-Mendoza et al. 2013). However, we have not been able to find a single example of a peach cultivar currently under commercial cultivation which contains beneficial genes from other species. Linkage drag, enhanced by suppression of recombination, low fertility, and the length of time usually required for recovering an elite genotype from an interspecific or wide cross have limited the adoption of this strategy in peach breeding programs.
An almond × peach progeny was developed at IRTA to obtain a high-density linkage map (Dirlewanger et al. 2004), taking advantage of its high polymorphism in contrast to the low segregation in the peach × peach crosses, facilitating map construction. This provided the scientific community with a common terminology and orientation for linkage groups and a large set of transferable markers, used as anchors for constructing other maps (Arús et al. 2012) and to align the physical map assembled for construction of the whole genome sequence (Verde et al. 2013). This map has been recently enhanced with information from a peach SNP chip, and nearly 2000 additional markers have been mapped to this and to an additional backcross (BC1) population from the same cross (Donoso et al. 2015). In this work, we analyzed these two interspecific populations to understand the genetic variability of 42 traits, exploring possible alleles from almond that could be introgressed into peach commercial cultivars. This information will be essential for the development of fast approaches for introgressing genes of interest from almond into the commercial peach gene pool.
Materials and methods
Plant material
Two interspecific populations (Donoso et al. 2015) were used. The first one was an F2 of N = 111 (T × E) obtained by selfing a hybrid individual (‘MB 1.37’) from a cross between almond ‘Texas’ and peach ‘Earlygold.’ The trees of T × E used for this research are at the IRTA Experimental Station of Lleida in Gimenells (Spain) grafted on ‘Garnem’ (Felipe 2009) rootstocks. The other was a BC1 (T1E) of 190 individuals, created from the cross ‘MB 1.37’ × ‘Earlygold.’ Original trees of T1E were planted on their own roots in Cabrils, and a replicate grafted on ‘Garnem’ was planted in the fields of Gimenells. Spacing within and between rows was 1.0 × 4.0 m for T1E in Cabrils, and 1.5 × 3.5 m for both populations in Gimenells. Standard agricultural practices were applied in both locations, but trees were not thinned out. Cabrils and Gimenells have contrasting climates: in Cabrils, located on the Mediterranean coast 25 km North of Barcelona, it is mild, whereas Gimenells, in central Catalonia, 150 km west of Barcelona, has a continental climate.
While all plants from T × E and T1E were used for mapping, some of them could not be phenotyped. This was because some plants died after DNA extraction for mapping (21 in T × E and 16 in T1E), not all plants were kept in both locations, and some characters could only be measured in part of the progenies (i.e., sterile plants did not produce fruit, precluding the analysis of fruit-related characters with them). The number of plants studied for each character, year and location is given in Supplementary Tables 1 and 2.
Phenotyping
Both populations were evaluated for 40 traits over 2 years (2012 and 2013), T × E in only one location (Gimenells) and T1E in both (Cabrils and Gimenells). In 2011, data were not available for blooming density and all leaf parameters in the T1E population of Cabrils (a total of 32 traits were measured), and only available for pistil length, fruit production, maturity date, and all fruit characters (21 traits in total) in both populations at Gimenells. The T1E population in Cabrils was evaluated for two additional characters: the juvenility period (2008–2013) and leaf fall date (2011–2013). The final set of 42 traits was divided into five categories as described below.
Flower traits
Flower type, the showy versus non-showy flower character, was described originally by Connors (1920) as a major gene (Sh/sh) in peach. Showy flowers are determined by the recessive allele (sh) and have large petals. Non-showy flowers have small petals, with anthers emerging from the corolla before full anthesis. Anther color, scored as anthocyanic anther or yellow anther in T × E, was described as a Mendelian trait (Ag/ag) in this population and mapped to linkage group 3 (G3) by Arús et al. (1994). Given that certain individuals initially scored as yellow had anthocyanic spots when observed with the binocular lens, all yellow phenotypes were reexamined with the binocular lens and re-scored as anthocyanic if these spots were visible. The yellow anther was determined by the recessive homozygote agag, whereas the presence of the Ag allele produced anthocyanic anthers. T1E individuals also segregated for anther color, although the phenotype had either a light anthocyanic coloration perceived as orange anthers, or a deep anthocyanic color perceived as red anthers. No plants had yellow anthers. We called this character AG2. Flower color (FC2) was classified as pink or pale pink. Pistil length (PIL) was scored as short (1), medium (2) or normal size (3). The blooming density (BD) character was scored visually using a scale from 1 to 5 (1 low; 2 low–medium; 3 medium; 4 medium–high; and 5 high).
Phenology traits
Beginning of shooting (BS) was scored as the number of Julian days at the moment when 5 % of shoots were in expansion. The beginning (BFT) and end (EFT) of flowering time were scored as the number of Julian days when 5 and 95 % of flowers were open, respectively. Flowering duration (FD) was calculated as EFT-BFT. Fruit production (FP) was scored on a scale from 1 to 4 (1 no fruits; 2 less than 10 fruits; 3 from 10 to 50 fruits; and 4 more than 50 fruits). Maturity date (MD) was scored as the number of Julian days when 50 % of the fruits were considered mature, based on visual color change and manual evaluation of firmness. The fruit development period (FDP) was the difference between scores of EFT and MD. Leaf fall (LF) was scored as the number of Julian days when 95 % of the leaves had dropped. The juvenility period (JUV) was scored as the number of years to produce the first fruits after germination.
Fruit traits
Fruit traits were evaluated in five mature fruits of each individual. Fruit type (ALF for almond fruit) was scored in the T × E population as peach or almond type, depending on the development of a fleshy mesocarp and a change in external and internal fruit color. In T1E, all individuals were peach type. Juiciness (JUI) was scored as the capacity of producing juice (1) or not (2). Blood flesh (BF2) was scored as presence (2) or absence (1) of red flesh color. The intensity of the red skin color (ISC) was scored from 1 to 4 (1 light red; 2 dark red; 3 dark violet red; and 4 violet). Fruit skin color was a qualitative estimation of the percentage of surface covered by anthocyanin coloration (PSC) according to the following scale: 1 = 0–25 %, 2 = 25–50 %, 3 = 50–75 % and 4 = 75–100 %. Soluble solid content (SSC) was scored as Brix degrees by applying a drop of fruit juice in a digital refractometer PAL-1 (Atago, Tokyo, Japan). For titratable acidity (TA) evaluation, two slices from each fruit were cut and stored at −20 °C prior to evaluation. TA was determined as g/l of malic acid by titrating 10 ml of distilled water and 10 ml of the unfrozen fruit juice or fruit paste with 0.1 N NaOH to pH 8.2. Fruit weight (FW) and stone weight (SW) were determined as the average of the five fruits scored, and flesh weight (FLW) as the difference FW-SW, all expressed in g. The following characteristics were measured to evaluate fruit and stone shape (see Fig. 1): fruit polar diameter (FPD), fruit cheek diameter (FCD), fruit suture diameter (FSD), stone polar diameter (SPD), stone cheek diameter (SCD) and stone suture diameter (SSD). Flesh diameters, flesh polar diameter (FLPD), flesh cheek diameter (FLCD) and flesh suture diameter (FLSD) were the difference between FPD and SPD, FCD and SCD, and FSD and SSD, respectively.
Scheme of the measurements taken for a, fruit and stone (FPD fruit polar diameter, FCD fruit cheek diameter, FSD fruit suture diameter, SPD stone polar diameter, SCD stone cheek diameter, SSD stone suture diameter), and b leaf dimensions (LL leaf length, LBL leaf blade length, LBW leaf blade width, PL petiole length)
Leaf traits
Eight leaves per tree were collected in July. To minimize leaf variation from each tree, leaves were collected from the middle of sun-exposed branches, with medium vigor and at 1.5–1.8 m high. Leaves were scanned and images saved as jpeg files and imported into Tomato Analyzer 3.0 for automated phenotypic measurements (http://www.oardc.ohio-state.edu/vanderknaap). The parameters analyzed were as follows: leaf perimeter (LP), leaf surface (LS), leaf blade width (LBW), leaf length (LL), leaf blade length (LBL) and, as the difference LL-LBL, petiole length (PL). All parameters were expressed in cm except for LS, expressed in cm2. Subsequently, the leaves were dried for 3 days at 60 °C and weighed to obtain the average leaf dry weight (LW). Figure 1 shows a sketch of the leaf dimensions measured. Chlorophyll content (CC) was measured with a SPAD-502 device (Konica Minolta, Osaka, Japan) that emits two wavelengths (650 and 940 nm), with the final measurement of chlorophyll content being the ratio between the two intensities of light after passing through each leaf.
Disease resistance
Powdery mildew resistance (VR3) was scored as the presence or absence of the disease in both populations by natural exposure. The trees were not treated during the years that this trait was evaluated (2011–2013). Trees with clear disease symptoms in at least 1 year were considered as susceptible.
Phenotypic data analysis
Statistical analyses were performed using JMP 8.0 software (SAS Institute, Cary, NC, USA). The distribution for each trait was represented in frequency histograms. Correlations between different traits and years were calculated using the Pearson correlation coefficient. The adjustment of the data to a normal distribution for each trait was tested with the Shapiro–Wilk test (Shapiro and Wilk 1965). The histograms were drawn using R 3.1.0 software (http://cran.r-project.org/bin/windows/base/).
Genetic linkage maps and QTL analysis
For linkage analysis, we used the data from the 9k International Peach SNP Consortium (IPSC) SNP Illumina Infinium chip (Verde et al. 2012) in 50 plants of the T × E population and 123 plants of the T1E population. In addition, we mapped 135 SSRs covering the whole genome in 111 T × E and 190 T1E plants (94 SSRs in common). Maps were constructed with these markers by Donoso et al. (2015). In summary, the T × E map contains 1948 markers and covers a total distance of 472.1 cM. The T1E population was used to construct two maps, one (the T1E map) with the markers heterozygous in the female parent, the MB1.37 hybrid plant, that contains 2031 markers and spans 370.1 cM, and the other (the E map) obtained with the markers heterozygous in the male ‘Earlygold’ parent with 1091 markers covering 520.4 cM.
Eight traits generated information on nine simple Mendelian factors that were added to the rest of the marker data files and mapped as individual genes using MapMaker v3.0 (Lander et al. 1987). These were flower color (Fc2), anther color (Ag and Ag2), juiciness (Jui), blood flesh (DBF2 for dominant blood flesh), flower shape (Sh), almond fruit type (Alf), maturity date (MD) and powdery mildew resistance (Vr3).
The remaining traits were subjected to QTL analysis using the interval mapping method with the MapQTL 4.0 software package (Van Ooijen et al. 2002). QTLs with a LOD ≥ 3.0 were declared significant. When a QTL with a LOD ≥ 2.5 was identified, all other years with LOD ≥ 3.0 were considered significant too. Maps and QTL positions were drawn using the MapChart 2.1 software (Voorrips 2002). We considered a QTL as consistent if it was detected every year in at least one of the locations, having overlapping confidence intervals (those established 1 LOD score unit below the maximum for each specific QTL). Given that certain traits were highly correlated, it was possible to group the traits studied in categories. Consistent QTLs detected in traits of a given category that mapped at overlapping positions and produced similar effects could be reasonably assumed to be the same, so we named them as major QTLs, allowing for a considerable reduction in the number of QTLs found.
Gene action was estimated in T × E following the guidelines of Tanksley (1993) with the ratio, d/a, between the additive, a = (A − B)/2, and dominance d = H − [(A + B)/2] effects, where H, A and B are the average phenotypic values of the heterozygous, almond homozygous and peach homozygous genotypes, respectively. Based on the d/a ratio, QTLs were classified as underdominant (d/a < −1.25; U), dominant for the peach allele (−1.25 ≤ d/a ≤ −0.75; DP), partially dominant for the peach allele (−0.75 ≤ d/a ≤ −0.25; PD), additive (−0.25 ≤ d/a ≤ 0.25; A), partially dominant for the almond allele (0.25 ≤ d/a ≤ 0.75; AD), dominant for the almond allele (0.75 ≤ d/a ≤ 1.25; DA) and overdominant (d/a > 1.25; O).
Results
Trait distributions and correlations
The T × E and T1E populations segregated for the 42 studied traits that were classified in five groups: five for flower, nine for phenology, 19 for fruit, eight for leaf and one for disease resistance. Five of these traits could be analyzed as qualitative, three as qualitative in one population and quantitative in the other (maturity date, juiciness and blood flesh), and the remaining 34 were considered only as quantitative. For four quantitative traits (pistil length, fruit production, blood flesh and juiciness) measured with a numeric score, the data for each year were identical and a single score per plant was used for the period analyzed. By its nature, the juvenility period for 2008–2013 consisted of a single score per plant. Distributions of these characters of parents and progenies for T × E and T1E are shown in Supplementary Tables 1 and 2 and Supplementary Figure 1.
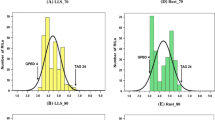
Variability for most traits was very high (see Fig. 2 for a sample of fruit traits), particularly in the T × E population. The hybrid individual MB1.37 had intermediate values between T and E in most cases. Transgressive segregation was observed in both populations for most traits, except for maturity date, fruit development period and fruit weight. Most fruit characters, except those related to juiciness, titratable acidity and skin color, had a normal distribution in both populations. All leaf characters were distributed normally in T × E, and only about half in T1E (18 of 34). On the other hand, pistil size, blooming density, most phenology traits, titratable acidity, intensity and percentage of skin color, and stone weight did not fit a normal distribution.
Correlations between years and locations for a given trait were usually positive and significant (P < 0.01). Exceptions were flowering duration in all years and locations in both populations, titratable acidity in 2011 in T × E, and chlorophyll content in 2012 in T1E (Supplementary Tables 3 and 4).
Several groups of quantitative characters were highly correlated, allowing us to divide the 37 characters studied into 18 categories. Six of these categories included several traits: shooting and flowering time (including BS, BFT and EFT); maturity time (MD and FDP); skin color (ISC and PSC); fruit dimensions (FW, FPD, FCD, FSD, FLW, FLPD, FLCD and FLSD); stone dimensions (SW, SPD, SCD and SSD) and leaf dimensions (LP, LL, LBW, LBL, LS and LW). The remaining twelve categories each had a single trait (PIL, BD, FP, LF, JUV, BF, JUI, FD, SSC, TA, PL and CC). Negative correlations were found in T × E between traits in the categories of maturity time and those of skin color and fruit dimensions. In T1E, skin color characters were negatively correlated with most other characters.
Mapping Mendelian traits
Eight of the traits studied were scored as qualitative, and nine Mendelian genes were mapped (Table 1): four (Fc2, Ag2, Jui and DBF2) in the T1E map, one (Sh) in the E map, three (Ag, Alf and MD) in the T × E map and one (Vr3) in both T1E and T × E. For one trait, anther color, we were able to extract information on two genes, one already described by Arús et al. (1994) mapped in T × E (Ag) and the other in T1E (Ag2). All traits had segregation ratios as expected under the single-gene hypothesis, except for Fc2, which had an excess of individuals with pink petals, and Vr3 (only in T × E) with an excess of resistant individuals. Six of these genes (Fc2, Ag2, DBF2, Jui, Alf and Vr3) were described and mapped for the first time. Details on their inheritance and position are given in Table 1.
Three of the six genes mapped for the first time determined color-related traits. Fc2 encodes for flower color, from white (Fc2Fc2), as in ‘Texas,’ to pale pink (Fc2fc2) and pink (fc2fc2), as in ‘Earlygold,’ and was located at the beginning of G4 in T1E. Flower color variability was also observed in the T × E population, but we were not able to identify a clear color pattern that allowed qualitative classification. A second anther color gene, Ag2, segregating only in T1E with orange (Ag2ag2) versus red (ag2ag2) phenotypes, was mapped on G1. The third gene, DBF2, determined the fruit blood flesh, with the dominant DBF2 allele from almond being responsible for red flesh color, mapped to the end of G1. Two more genes encoded fruit quality characters. One, the Jui gene, determines juiciness, with juicy fruit being homozygous for the recessive jui allele, from peach, and the Jui- genotype, non-juicy. It mapped to the distal position of G1. The other gene, Alf, determines the capability of developing a thick fruit mesocarp, as in peach, conferred by the dominant Alf allele from peach, whereas recessive homozygotes, alfalf, have an almond-like fruit. This gene segregated only in T × E, mapping on G4 at the same position as MD. Finally, the gene for resistance to powdery mildew (Vr3) was mapped in both populations at a similar position on G2. Although this trait was evaluated using the natural inoculum in the field, data were always consistent between the different years evaluated.
Genetic analysis of quantitative characters
Taking into consideration all the years, locations and the three maps analyzed, a total of 155 QTLs were detected: 100 in T1E, six in E and 49 in T × E for the 37 quantitative traits studied (Supplementary Table 5 and supplementary Figure 2). QTLs were identified for all traits, varying between one and four QTLs per trait. We found 61 consistent QTLs (39.4 % of the total): 42 in the T1E map, two in the E map and 17 in the T × E map (Supplementary Table 6). For some traits, such as maturity date and petiole length, most of the QTLs detected were consistent, while in others, such as SSC, TA and flowering duration, none of the QTLs detected was consistent. Considering that the consistent QTLs with similar positions and effects for the trait categories mentioned previously were the same, we could reduce their number to 32 major QTLs (Table 2; Fig. 3). A description of these is given in the following paragraphs.
Map with the positions of the major genes and QTLs mapped in this work. The map used was that of the T × E population, and the positions of major genes or QTLs mapped on the other two maps (T1E and E) are inferred from that of common markers. Only the 32 major QTLs were mapped, and its position was that of the consistent QTL within the highest LOD, when more that one consistent QTL was included in the major QTL. Colors of the bars of QTLs or gene names were as follows: pink flower, blue phenology, red fruit, green leaf-related traits and brown the powdery mildew resistance gene. (Color figure online)
For pistil length, a major QTL explaining a large percentage (R 2 = 74.4 %) of the phenotypic variance was detected at the end of G6 in T × E. In T1E, two QTLs were detected on G1 and G4. For blooming density, a single major QTL was detected in T1E, at the end of G1, and only in Gimenells, with R 2 = 23.8–25.7 %.
For shooting and blooming time, six major QTLs were identified, one on each of G1, G6, G7 and G8, and two on G2. The one on G1 was in common between T × E and T1E, and the rest were specific of T × E (G6), E (G7) and T1E (G8 and both QTLs on G2). A large QTL for fruit production was detected in the central region of G6 in T × E and T1E (R 2 = 45.7 and 33.3 %, respectively). This QTL colocalized with a QTL for juvenility in T1E (R 2 = 13.3 %). A QTL for leaf fall date was detected at the beginning of G8 in T1E. A single major QTL on G4 (R 2 = 23.8–57.9 %) was identified for maturity date characters. This QTL peaked at the same region where the MD gene was located in T × E. The presence of the almond allele at this locus increases the MD an average of 32 days in T1E and 42 days in T × E.
A large QTL for juiciness (R 2 = 31.8 %) was found in T × E at the end of G1, colocating with the position of the Jui gene mapped in T1E. For the blood flesh trait, one QTL was detected in the central region of G3 in T × E (qBF3), colocating with the position of the anther color gene (Ag), explaining 50.7 % of the phenotypic variance. Skin color traits detected two major QTLs on G1 and G4 in T1E and one on G3 in T × E.
Fruit dimensions were explained in T × E by a major QTL found in the central region of G4, at the same position as the MD and Alf genes, accounting for much of their phenotypic variability (35.8–77.5 %). In this population, individuals heterozygous at this locus had larger fruits than those homozygous for the peach and the almond alleles. A different picture occurs in T1E, where ALF did not segregate and two QTLs were detected, one at the beginning of G6, and another in the middle of G7, explaining around 20 % of their phenotypic variance. For the traits of stone dimensions, a major QTL was detected in T1E, in the central part of G6, explaining between 28.1 and 46.0 % of the phenotypic variance.
Four major QTLs were identified for leaf dimensions (on G1, G6, G7 and G8). One of them was associated with leaf width (G6) and another with leaf surface (G7). Petiole length appeared to have a different genetic determinism, with four QTLs on G5, where no QTLs were found for leaf characters, and three more (on G6, G7 and G8) at different positions to those found for leaves. For chlorophyll content, a major QTL was identified in TIE, on G6, (qCC6).
Discussion
We mapped nine major genes and 32 major QTLs using two almond × peach interspecific populations. From the QTLs identified, 26 were in the BC1 population T1E (25 mapped in the map of the almond × peach interspecific hybrid used as female parent and one in the peach ‘Earlygold’ male parent), and ten in the F2 progeny T × E, four in common with T1E. We expected more segregating traits in T × E than in T1E, because those with a dominant gene action in the peach parent would not be detected in the BC1 population, and more QTLs in common between T × E and T1E. The common QTLs were for fruit production (qFP6), flowering time (qSF1), and two for petiole length (qPL5 and qPL8). We could add two more that were mapped as quantitative in one population and as qualitative in the other: maturity date (qMD4, MD) and juiciness (qJUI1, Jui). These results can be explained by the larger size of T1E, resulting in greater statistical power to identify QTLs with low effects (Beavis 1998), or by the more uniform background of the BC1, with less interactions between genes of different parts of the genome affecting the same character, allowing more accurate phenotyping. One example of the latter is the group of traits in the fruit dimensions category that, in the case of T × E, were strongly affected by individuals with almond-like fruit type that were not present in the T1E progeny. In T × E, all these traits identified only one QTL on G4 at the position of the Alf gene, whereas no QTL was detected on G4 in the T1E progeny, although two were identified on G6 and G7.
Only one major QTL was detected with the E map. We attribute this to the lower level of genetic variability within peach, with about half of the ‘Earlygold’ genome identical by descent (Donoso et al. 2015), and by the fact that more subtle phenotypic effects of alleles segregating in ‘Earlygold’ could often be masked by greater effects from the almond alleles.
The analysis of T1E from two locations was an opportunity to detect possible genotype × environmental interactions. We identified six consistent QTLs that were only found in one of the locations (see Supplementary Table 6): two for beginning of flowering (qBFT8) and leaf length (qLL1) were only present in Cabrils; four, determining blooming density (qBD1), fruit weight (qFW6), flesh weight (qFLW7) and fruit cheek diameter (qFCD7), were detected only in Gimenells. For qBFT8, qBD1 and qLL1, this may respond to the important climatic differences between the two locations. The traits related to fruit dimensions may be associated with the culture practices, such that fruits from plants growing in Gimenells under standard agronomic conditions were larger than those from plants in Cabrils grown on their own roots and with less spacing between trees.
Major genes
Interspecific crosses allow the analysis of traits with contrasting phenotypes in the species involved. Some of these traits have a simple inheritance and allow the identification of major genes or major QTLs that explain much of the observed variability. In our almond × peach crosses, we have been able to map eleven major genes, nine described here and two previously published (Donoso et al. 2015), corresponding to two male fertility restorer genes on G2 and G6 (Rf1 and Rf2). In addition, several traits resolved in QTLs explaining more than 50 % of the phenotypic variation, such as pistil length in G6 of T × E or blood flesh in G3 of T1E, suggesting that a large proportion of the variability detected was controlled by genes/QTLs of major effects. Most of these major genes (nine of 11) had not been mapped before in other peach populations, and for the other two, Sh (Ogundiwin et al. 2009) and MD (Eduardo et al. 2011), our data corroborated their positions on G8 and G4, respectively.
Two of the major genes discovered, fruit type (almond vs. peach; Alf) and juiciness (juicy vs. non-juicy; Jui), are key distinctive features between the fruit of peach and almond. A third locus, the gametophytic self-incompatibility, operative in almond but not in peach, is a major determinant of another crucial difference between almond and peach: their extremely diverse pattern of genetic variability, very low in peach and enormously high in almond (Mnejja et al. 2010). The fact that such a simple genetic basis explains essential differences between these two crops confirms their close genetic origin, suggesting that they may have been domesticated from close relatives or even from the same species.
Some of the major genes found in these almond × peach progenies for traits such as fruit flesh and flower anthocyanic color, or powdery mildew resistance, were located, interestingly, at chromosomal positions different from genes described previously in other populations, mostly peach × peach, for the same characters. Two major genes for fruit flesh color have been described, Bf on G4 by Gillen and Bliss (2005), and DBF on G5 by Shen et al. (2013), whereas two loci found in our materials for the same trait (DBF2 and qBF3) mapped elsewhere (G1 and G3, respectively). A gene for flower color (Fc) was mapped on G3 by Yamamoto et al. (2001), while we found a gene for the same trait on G4 (Fc2). For powdery mildew resistance, the Vr3 gene that we mapped to G2 had a different genome position to those described previously, one on G6 (Vr2; Pascal et al. 2010) and several QTLs on G7 (Verde et al. 2002; Pacheco et al. 2009), G6 and G8 (Foulongne et al. 2003). These results indicate that the variability supplied by almond is often in different genes than already described. This enhances its interest as a source of novel variability that can be combined to that already available to produce cultivars with added value. One obvious example is that of resistance to powdery mildew on G2 that can be pyramided with known others to obtain cultivars with broader and more durable resistance.
QTLs
Flower traits
A QTL for pistil length was identified in T × E on G6. Given that most plants with short pistils were unable to bear fruit, this trait was highly correlated with fruit production in T × E and a major QTL for this character was detected in the same region. The same locus on G6 contains one or more genes that affect fruit shape (round vs. flat), aborting fruit, pistil length and fruit production (Dirlewanger et al. 2006; Picañol et al. 2013). In T1E, a QTL for fruit production was also detected in the same region of G6, but QTLs for pistil length mapped to G2 and G4, suggesting that the gene(s) involved in pistil length and those determining productivity at the G6 region are not the same.
For blooming density, we identified a major QTL on G1, where the peach allele decreased flower density. This is an important trait in Prunus, as it determines the need for flower or fruit thinning during the growing season, one of the most expensive operations of peach cultivation.
Phenology traits
Six consistent QTLs were detected for traits related with the time of shooting and blooming, which are highly heritable traits and appear to be related, as suggested by the high correlations that we observed between them, and by the fact that some of the QTLs (on G7 and G8) appeared to be in common. However, some authors (Kester et al. 1973; Dicenta et al. 1993) have reported low correlations between shooting and blooming times, indicating that specific elements of their inheritance are also important. Our data support a complex genetics for these traits, with many genes involved and different sets of them, partly overlapping between traits, segregating in each mapping population. The same pattern has been found in peach and other Prunus crops (Wang et al. 2000; Quilot et al. 2004; Sánchez-Pérez et al. 2012; Romeu et al. 2014; Socquet-Juglard et al. 2013; Dirlewanger et al. 2012).
MD was mapped as a single gene in T × E and as a major QTL in T1E. This locus has already been identified as the major factor determining this character in peach, apricot and sweet cherry (Dirlewanger et al. 2012), suggesting a common fruit maturation control mechanism.
We scored the juvenility trait as the number of years required by each T1E plant to produce fruit for the first time, with values that ranged from three to six, and identified a QTL (qJUV1) that explained 13.3 % of the phenotypic variance, with the peach allele increasing the juvenility period. No previous information exists on the genetics of this character in Prunus, but in citrus, Raga et al. (2012) detected four QTLs related to juvenility that jointly explained 39.2 % of the phenotypic variance. Knowledge of the inheritance for this trait may be helpful to breed lines with shorter intergeneration periods that would speed up the breeding process.
Fruit traits
The complex patterns of the anthocyanic pigment coloration in plants appear to be determined by allelic variation in the enzymes of the anthocyanin biosynthetic pathway and their interaction with various transcription factors. This results in a diverse set of phenotypes in various organs and life cycle stages or as a response to environmental change (Rahim et al. 2014; Zhou et al. 2014; Espley et al. 2009). The pattern that we found in our research is compatible with this scenario, where map positions of phenotypic variants and these genes colocate. One of the colocations that we identified was on G3, with three loci, Ag, qPSC3 and qBF3, determining color in anthers, fruit skin and fruit flesh, respectively. QTLs have also been found in this region for fruit red blush in peach (Frett et al. 2014) and for skin and flesh color in sweet cherry (Sooriyapathirana et al. 2010). This region contains a cluster with three genes of the MYB transcription factor family (Rahim et al. 2014) that appear evident candidates for the observed variability. A homolog of one of these genes in apple (MdMYB10) is responsible for its red fruit flesh color (Espley et al. 2009). Another region with an accumulation of color genes is at the end of G1, where DBF2 and qSC1 map and where a major gene for flower color (B) has also been located (Dirlewanger et al. 2004). This region encompasses a gene coding for one of the key enzymes of the anthocyanin pathway: a flavonol synthase, FLS1; ppb018307 m.
For fruit and stone dimensions, only one major QTL at the Alf position on G4 was identified in T × E. The major effects of this QTL probably masked other QTLs with smaller effects. For T1E, all plants are peach type (Alf-), and two interesting QTLs were identified. One was on G7, explaining a decrease in flesh weight (21–25 g) produced by the almond allele. The other affected stone parameters, where the almond allele of a QTL on G6 (qSD6) determined an increase in stone weight (3–4 g). A QTL at the beginning of G6 (qFW6) was found only for fruit weight, with heterozygous plants having heavier fruit (25–34 g) than those homozygous for the peach allele. This QTL was significant for flesh weight (increasing the almond allele 27 g) only in two of the 3 years studied (see Supplementary Table 5), so it was not considered consistent. The existence of qFW6 indicates that it is possible to find genes in the almond that can be used for increasing fruit size, one of the classical objectives of peach breeding. QTLs for FW on G6 have been described in cherry (Zhang et al. 2010; Rosyara et al. 2013), peach (Dirlewanger et al. 1999; Yamamoto et al. 2001; Eduardo et al. 2011) and peach × P. davidiana (Quilot et al. 2004). In the latter work, a QTL on G6 was also found for stone size. Overall, our results suggest a different genetic basis for stone versus flesh dimension traits, which would make it possible to select for increased flesh weight and decreased seed weight in peach, or the opposite in almond.
Leaf traits
The QTLs identified for leaf parameters suggest that each acts on a different aspect of this trait. A QTL on G1 explains, in part, the variation in the overall leaf size and is detected by all leaf measurements (perimeter, surface, width, length and weight). Another QTL, located on G8, is specific to leaf length and is detected in all leaf traits except in width and weight, with the almond allele producing leaf elongation. A QTL affecting only the leaf width trait was detected on G6, the almond allele producing an increase in this parameter. For leaf surface, we were able to identify a consistent QTL on G7 that did not appear in any other trait. Our interpretation is that leaf surface is a trait that better integrates certain aspects of the leaf shape, allowing the detection of an additional QTL. The only reported result for leaf dimensions in peach is a major gene, Nl, for leaf shape (narrow vs. wide) identified by Yamamoto et al. (2001). This locus maps to a genomic region coincident with the QTL for leaf width located here on G6, suggesting that they could correspond to the same locus. The inheritance of petiole length, determined by four QTLs located on G5, G6, G7 and G8, with the peach allele producing a decrease in petiole length in all cases, appears to identify a gene network completely different from that of the other leaf dimensions.
Conclusions
In this paper, we provide a first survey on the almond variability available for peach breeding. We described nine major genes and many QTLs explaining large fractions of the phenotypic variability. Most of these genes are at different genomic positions to others already described for similar traits in peach, meaning that they supply new variability that can be combined with that already existing. Recent work (Fresnedo-Ramírez et al. 2015) on progenies between peach and related species (almond and other peach close relatives) found six consistent QTLs for some of the traits studied here. They only had one QTL (that of MD on G4) in common, suggesting the existence of an immense gene pool in these species, of which the results from this research have unveiled only a minimal part. Some of these genes/QTLs have immediate applications for plant breeding: Resistance to powdery mildew (Vr3), the blood flesh (DBF2) locus, the major QTLs for skin color in G1 and the QTL on G6 that increases fruit weight are examples of almond alleles that produce traits of interest in peach and could be introgressed in its genome.
The presence of major genes for certain characters made it difficult to study other genes or QTLs for similar traits that had weaker effects. We have seen that our ability to detect QTLs improved in T1E, where the almond alleles were expressed in a more uniform peach genetic background, but at the cost of not being able to detect recessive almond alleles. One way for improvement would be using more numerous progenies, requiring a much larger expenditure in time and cost in fruit trees than in herbaceous species, or by developing specific populations such as introgression line collections (Eshed and Zamir 1994) that allow for a more accurate analysis of the almond variability, minimizing background effects. This is our next step from the materials that we obtained in ‘Texas’ × ‘Earlygold’ crosses, which in addition to providing a high-quality resource for genetic analysis, may prove an efficient first step toward introgression of almond alleles into peach. The development of marker-based methods to make introgression faster is another way of fostering the use of genes from other Prunus into peach, expanding its impoverished gene pool and making it possible to reach new goals on fruit quality, shelf life and stress resistance, of interest for both growers and consumers.
References
Arús P, Messeguer R, Viruel M, Tobutt K, Dirlewanger E, Santi F, Quarta R, Ritter E (1994) The European Prunus mapping project: progress in the almond linkage map. Euphytica 77(1–2):97–100
Arús P, Verde I, Sosinski B, Zhebentyayeva T, Abbott AG (2012) The peach genome. Tree Genet Genomes 8:531–547
Beavis W (1998) QTL analyses: power, precision, and accuracy. In: Paterson AH (ed) Molecular dissection of complex traits. CRC Press, Boca Raton, pp 145–162
Byrne DH, Raseira MB, Bassi D, Piagnani MC, Gasic K, Reighard GL, Moreno MA, Pérez S (2012) Peach. In: Badenes ML, Byrne DH (eds) Fruit breeding (handbook of plant breeding). Springer, New York, pp 513–569
Cao K, Zheng Z, Wang L et al (2014) Comparative population genomics reveals the domestication history of the peach, Prunus persica, and human influences on perennial fruit crops. Genome Biol 15:415
Carrillo-Mendoza O, Chaparro JX, Williamson J (2013) Branching and blind node incidence in interspecific backcross families of peach. HortScience 48:1119–1124
Claverie M, Bosselut N, Lecouls AC, Voisin R, Lafargue B, Poizat C, Kleinhentz M, Laigret F, Dirlewanger E, Esmenjaud D (2004) Location of independent root-knot nematode resistance genes in plum and peach. Theor Appl Genet 108:765–773
Connors CH (1920) Some notes on the inheritance of unit characters in the peach. Proc Am Soc Hortic Sci 16:24–36
Dicenta F, García JE, Carbonell EA (1993) Heritability of flowering, productivity and maturity in almond. J Hortic Sci 68:113–120
Dirlewanger E, Moing A, Rothan C, Svanella L, Pronier V, Guye A, Plomion C, Monet R (1999) Mapping QTLs controlling fruit quality in peach [Prunus persica (L.) Batsch]. Theor Appl Genet 98:18–31
Dirlewanger E, Graziano E, Joobeur T, Garriga-Calderé F, Cosson P, Howad W, Arús P (2004) Comparative mapping and marker-assisted selection in Rosaceae fruit crops. Proc Natl Acad Sci USA 101:9891–9896
Dirlewanger E, Cosson P, Boudehri K, Renaud C, Capdeville G, Tauzin Y, Laigret F, Moing A (2006) Development of a second-generation genetic linkage map for peach [Prunus persica (L.) Batsch] and characterization of morphological traits affecting flower and fruit. Tree Genet Genomes 3:1–13
Dirlewanger E, Quero-Garcia J, Le Dantec L et al (2012) Comparison of the genetic determinism of two key phenological traits, flowering and maturity dates, in three Prunus species: peach, apricot and sweet cherry. Heredity 109:280–292
Donoso JM, Eduardo I, Picañol R, Batlle I, Howad W, Aranzana MJ, Arús P (2015) High-density mapping suggests cytoplasmic male sterility with two restorer genes in almond × peach progenies. Hortic Res 2:15016
Eduardo I, Pacheco I, Chietera G, Bassi D, Pozzi C, Vecchietti A, Rossini L (2011) QTL analysis of fruit quality traits in two peach intraspecific populations and importance of maturity date pleiotropic effect. Tree Genet Genomes 7:323–335
Eshed Y, Zamir D (1994) A genomic library of Lycopersicon pennellii in L. esculentum: a tool for fine mapping of genes. Euphytica 79:175–179
Espley RV, Brendolise C, Chagné D et al (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21:168–183
Felipe AJ (2009) ‘Felinem, ‘Garnem’ and ‘Monegro’ almond × peach hybrid rootstocks. HortSci 44:196–197
Foulongne M, Pascal T, Pfeiffer F, Kervella J (2003) QTLs for powdery mildew resistance in peach × Prunus davidiana crosses: consistency across generations and environments. Mol Breed 12:33–50
Fresnedo-Ramírez J, Bink MCAM, van de Weg E, Famula TR, Crisosto CH, Frett TJ, Gasic K, Peace CP, Gradziel TM (2015) QTL mapping of pomological traits in peach and related species breeding germplasm. Mol Breeding 35:166
Frett TJ, Reighard GL, Okie WR, Gasic K (2014) Mapping quantitative trait loci associated with blush in peach [Prunus persica (L.) Batsch]. Tree Genet Genomes 10:367–381
Gillen AM, Bliss FA (2005) Identification and mapping of markers linked to the Mi gene for root-not nematode resistance in peach. J Am Soc Hortic Sci 130(1):24–33
Gradziel TM (2003) Interspecific hybridizations and subsequent gene introgression within Prunus subgenus Amygdalus. Acta Hortic 622:249–255
Kester DE, Raddi P, Asay R (1973) Correlation among chilling requirements for germination, blooming and leafing in almond (Prunus amygdalus Batsch). Genetics 74:s135
Lander E, Green P, Abrahamson J, Barlow A, Daly M, Lincoln S, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Li XW, Meng XQ, Jia HJ et al (2013) Peach genetic resources: diversity, population structure and linkage disequilibrium. BMC Genet 14:84
Micheletti D, Dettori MT, Micali S et al (2015) Whole-genome analysis of diversity and SNP-major gene association in Peach Germplasm. PLoS ONE 10(9):e0136803
Mnejja M, Garcia-Mas J, Audergon JM, Arús P (2010) Prunus microsatellite marker transferability across rosaceous crops. Tree Genet Genomes 6:689–700
Ogundiwin EA, Peace CP, Gradziel TM, Parfitt DE, Bliss FA, Crisosto CH (2009) A fruit quality gene map of Prunus. BMC Genom 10:587
Pacheco I, Eduardo I, Rossini L, Vecchieti A, Bassi D (2009) QTL mapping for peach (Prunus persica (L.) Batsch) resistance to powdery mildew and brown rot. In: Proceedings of the 53rd Italian Society of Agricultural Genetics Annual Congress. Torino, Italy
Pascal T, Pfeiffer F, Kervella J (2010) Powdery mildew resistance in the peach cultivar Pamirskij 5 is genetically linked with the Gr gene for leaf color. HortScience 45:150–152
Picañol R, Eduardo I, Aranzana MJ, Howad W, Batlle I, Iglesias I, Alonso JM, Arús P (2013) Combining linkage and association mapping to search for markers linked to the flat fruit character in peach. Euphytica 190:279–288
Quilot B, Wu BH, Kervella J, GénardM FoulongneM, Moreau K (2004) QTL analysis of quality traits in an advanced backcross between Prunus persica cultivars and the wild relative species P. davidiana. Theor Appl Genet 109:884–897
Raga V, Bernet GP, Carbonell EA, Asins MJ (2012) Segregation and linkage analyses in two complex populations derived from the citrus rootstock Cleopatra mandarin. Inheritance of seed reproductive traits. Tree Genet Genomes 8:1061–1071
Rahim MA, Busatto N, Trainotti L (2014) Regulation of anthocyanin biosynthesis in peach fruits. Planta 240:913–929
Romeu JF, Monforte AJ, Sánchez G, Granell A, García-Brunton J, Badenes ML, Ríos G (2014) Quantitative trait loci affecting reproductive phenology in peach. BMC Plant Biol 14:52–68
Rosyara UR, Bink MCAM, van de Weg E, Zhang GR, Wang DC, Sebolt A, Dirlewanger E, Quero-Garcia J, Schuster M, Iezzoni AF (2013) Fruit size QTL identification and the prediction of parental QTL genotypes and breeding values in multiple pedigreed populations of sweet cherry. Mol Breeding 32(4):875–887
Rubio M, Pascal T, Bachellez A, Lambert P (2010) Quantitative trait loci analysis of Plum pox virus resistance in Prunus davidiana P1908: new insights on the organization of genomic resistance regions. Tree Genet Genomes 6:291–304
Sánchez-Pérez R, Dicenta F, Martínez-Gómez P (2012) Inheritance of chilling and heat requirements for flowering in almond and QTL analysis. Tree Genet Genomes 8:379–389
Sauge MH, Lambert P, Pascal T (2012) Co-localisation of host plant resistance QTLs affecting the performance and feeding behaviour of the aphid Myzus persicae in the peach tree. Heredity 108:292–301
Scorza R, Mehlenbacher SA, Lightner GW (1985) Inbreeding and coancestry of freestone peach cultivars of the eastern United States and implications for peach germplasm improvement. J Am Soc Hortic Sci 110:547–552
Shapiro SS, Wilk MB (1965) An analysis of variance test for normality (complete samples). Biometrika 52(3–4):591–611
Shen Z, Confolent C, Lambert P, Poëssel J-L, Quilot-Turion B, Yu M, Ma R, Pascal T (2013) Characterization and genetic mapping of a new blood-flesh trait controlled by the single dominant locus DBF in peach. Tree Genet Genomes 9:1435–1446
Shulaev V, Korban SS, Sosinski B et al (2008) Multiple models for Rosaceae genomic. Plant Physiol 147:985–1003
Socquet-Juglard D, Christen D, Devènes G, Gessler C, Duffy B, Patocchi A (2013) Mapping architectural, phenological, and fruit quality QTLs in apricot. Plant Mol Biol Rep 31:387–397
Sooriyapathirana SS, Khan A, Sebolt AM, Wang D, Bushakra JM, Lin-Wang K, Allan AC, Gardiner SE, Chagné D, Iezzoni AF (2010) QTL analysis and candidate gene mapping for skin and flesh color in sweet cherry fruit (Prunus avium L.). Tree Genet Genomes 6:821–832
Tanksley SD (1993) Mapping polygenes. Ann Rev Genet 27:205–233
Van Ooijen JW, Boer MP, Jansen RC, Maliepaard C (2002) MapQTL® 4.0. Software for the calculation of QTL positions on genetic maps. Plant Research International, Wageningen
Verde I, Quarta R, Cerdrola C, Dettori MT (2002) QTL analysis of agronomic traits in a BC1 peach population. Acta Hortic 592:291–297
Verde I, Bassil N, Scalabrin S et al (2012) Development and evaluation of a 9K SNP array for peach by internationally coordinated SNP detection and validation in breeding germplasm. PLoS ONE 7:e35668
Verde I, Abbott AG, Scalabrin S et al (2013) The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet 45:487–494
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93(1):77–78
Wang D, Karle R, Iezzoni AF (2000) QTL analysis of flower and fruit traits in sour cherry. Theor Appl Genet 100:535–544
Yamamoto T, Shimada T, Imai T, Yaegaki H, Haji T, Matsuta N, Yamaguchi M, Hayashi T (2001) Characterization of morphological traits based on a genetic linkage map in peach. Breed Sci 51:271–278
Zhang JB, Sebolt AM, Wang D, Bink M, Olmstead JW, Iezzoni AF (2010) Fruit size QTL analysis of an F1 population derived from a cross between a domesticated sweet cherry cultivar and a wild forest sweet cherry. Tree Genet Genomes 6:25–36
Zhou Y, Zhou H, Lin-Wang K, Vimolmangkang S, Espley RV, Wang L, Allan AC, Han YP (2014) Transcriptome analysis and transient transformation suggest an ancient duplicated MYB transcription factor as a candidate gene for leaf red coloration in peach. BMC Plant Biol 14:388
Acknowledgments
Funding of this research was granted in part by the Spanish Ministry of Economy and Knowledge (Project AGL2012-40228). We thank C. Fontich and A. Ortigosa (IRTA) for maintenance of the plant populations in Lleida and Cabrils, respectively.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Donoso, J.M., Picañol, R., Serra, O. et al. Exploring almond genetic variability useful for peach improvement: mapping major genes and QTLs in two interspecific almond × peach populations. Mol Breeding 36, 16 (2016). https://doi.org/10.1007/s11032-016-0441-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-016-0441-7