Abstract
Cucumber is a typical monoecious having separate male and female flowers on the same plant, and sex expression in cucumber is mainly determined by three major genes: F/f, M/m and A/a. Gynoecy plays an important role in cucumber hybrid breeding, and the use of gynoecious lines as maternal parents ensures high productivity. This study aimed to identify a co-dominant marker linked to Female (F) locus to distinguish homozygous and heterozygous gynoecious lines for efficient selection of gynoecy in cucumber breeding. Firstly, we analyzed a 50-kb genomic sequence of five gynoecious and five monoecious inbred lines to detect the polymorphism linked to F locus. A pair of specific primers, Cs-BCAT-F/Cs-BCAT-R, based on an In/Del polymorphism at 3′-UTR of branched-chain amino acid transaminase (BCAT) gene was designed to examine the polymorphism in gynoecious and monoecious parents, their F 1 hybrids and two F 2 segregating populations. The PCR fragments amplified with Cs-BCAT-F/Cs-BCAT-R co-segregated with sexual phenotypes in the F 2 populations, behaving as a co-dominant marker with two alleles: F (gynoecious) and f (monoecious). When we further verified the consistency of the co-dominant marker using additional 55 diverse inbred lines, it can explain ~90 % of accessions. Linkage analysis also showed that the gene which enhances the number of female flowers co-segregated with the co-dominant marker on the long arm of chromosome 6. This is the first report for the development of F locus-specific co-dominant marker which can distinguish perfectly homozygous and heterozygous gynoecious, and it could be used in marker-assisted selection in cucumber breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cucumber (Cucumis sativus L.) is a typical monoecious plant which composed of separate male and female flowers on the same plant. In addition, cucumber has additional three different sex types of flower; gynoecious, andromonoecious and hermaphroditic. Sex expression in cucumber is controlled by combinatory effect of genetic, environmental and hormonal factors (Malepszy and Niemirowicz-Szczytt 1991; Perl-Treves 1999). Inheritance and molecular mechanisms of sex expression in cucumber have been well characterized. Sex expression of cucumber flowers have been mainly determined by three sex-determination genes; a dominant Female (F) locus, and two other recessive loci andromonoecious (m, staminate and hermaphrodite flowers) and androecious (a, staminate flowers).
Female sex expression (gynoecy) plays an important role in cucumber breeding program because the commercial F 1 hybrid seeds were generated by crossing two different unisexual cucumber breeding lines. Therefore, selection of gynoecious cucumber lines used for maternal parents is the essential process in cucumber breeding. Traditional selection of gynoecious cucumber line has been based on observation of flower sex expression in breeding fields. The traditional method has several limitations in terms of selection accuracy, early identification and maintenance. Besides, unfavorable environmental conditions can reduce the selection accuracy under field conditions since sex expression in cucumber is affected by environmental conditions. Therefore molecular selection of gynoecy becomes essential to improve the efficiency and precision of conventional cucumber breeding.
Gynoecy is regulated by a single dominant gene, Female (F) (Shifriss 1961; Kubicki 1969), and the F locus encodes an additional copy of 1-aminocyclopropane-1-carboxylic acid synthase (ACS) gene, Cs-ACS1G, a key regulator of ethylene biosynthesis (Trebitsh et al. 1997). Cs-ACS1G appears to be the result of gene duplication and recombination between Cs-ACS1 and Cs-BCAT gene (Mibus and Tatlioglu 2004; Knopf and Trebitsh 2006), and the cis-regulatory elements needed for cucumber female-specific expression might be located in the distal promoter region of Cs-ACS1G (Trebitsh et al. 1997; Wu et al. 2012).
To date, it has been considered that structural variation of cucumber genome control the female sex expression. The recent 30.2-kb duplication causes a copy number variation involving four genes; Cs-ACS1, one unknown gene, a truncated MYB transcription factor and part of Cs-BCAT gene that defines the F locus (Zhang et al. 2015). Therefore, PCR amplicon could detect only gynoecious by using the two primer sequences spanning the breakpoint of the duplicated region. The molecular markers linked to F locus have been previously reported; such as the primer pair P2S/P3A covering the full length-specific genomic sequence of Cs-ACS1G (Knopf and Trebitsh 2006), the primer pair amplified the upstream of Cs-ACS1G gene covering the recombination event between Cs-ACS1 and Cs-BCAT genes (Wu et al. 2012), and the marker Primer F/Primer R spanning the breakpoint of the 30.2-kb duplicated region which defines the F locus (Zhang et al. 2015). Although those molecular markers could identify gynoecious lines, dominant characteristics of this marker could not distinguish homozygous and heterozygous gynoecious lines. Since heterozygous gynoecious lines could exhibit a less stable gynoecious phenotype compared with homozygous lines, it still needs the updated molecular marker to overcome this troublesome. Currently, some SSR markers linked to gynoecious locus in cucumber have been reported (Zhou et al. 2013). However, the applicability of these markers is still limited because there were several genetic distances between the markers and the F locus. Therefore, we analyzed the 50-kb genomic sequence including the 30.2-kb duplication of monoecious and gynoecious lines to develop F locus-specific co-dominant markers.
In this paper, we identify a new co-dominant molecular maker which can distinguish homozygous and heterozygous gynoecious cucumber lines. We also exploit the sequence polymorphism of gynoecious and monoecious lines in the duplicated region which defines the F locus. For the verification of F locus-specific identification, we also mapped the co-dominant marker onto the F 2 mapping population and screened the diverse cucumber accessions.
Materials and methods
Plant materials and growth conditions
Two Korean gynoecious lines of cucumber (C. sativus L.) ‘WJE F10’ and ‘WJE F11’ were used as female parents, and the monoecious line ‘WNE F8’ was used as a pollinator to generate F 2 segregating populations. The F 1 hybrids that resulted from each cross-combination were self-pollinated in summer 2014. Both parents, their F 1 hybrids and two F 2 populations consisting of 160 individuals derived from ‘WJE F11’ and ‘WNE F8’ and 60 individuals derived from ‘WJE F10’ and ‘WNE F8’ were cultivated in summer 2015 in a greenhouse in Sejong University, Seoul, South Korea. All plant materials were grown in pea bags with drip irrigation of nutrient solution, following plant protection practices as necessary. Furthermore, 55 accessions of diverse inbred lines with different sexual phenotypes from different origins (Table S1) were cultivated in a greenhouse in Sejong University for five continuous years from 2010 to 2014 to determine their sexual phenotypes.
Evaluation of sex expression
The sexual phenotype of individual plant was determined by recording the sex type of the first 25 nodes along the main stem in summer season from early June to late July. The plants with female flowers at every node were scored as gynoecious and those which bear male flowers at early 3–5 nodes and female flowers at all the remaining nodes were scored as subgynoecious, whereas the plants which produced both male and female flowers were scored as monoecious.
Whole genome sequencing
Genomic DNA from four gynoecious lines (TH169, WDSGD, WFEJ and WJE F11) and three monoecious lines (TH118FLM, WMEJ and WNE F8) was isolated from young leaves using DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). A library of ~280-bp insert size was constructed at Theragen BiO Institute (TBI, TheragenEtex, Korea) and pair-end sequenced (2 × 100 bp) on an Illumina HiSeq2000 using HiSeq Sequencing Kits. Raw pair-end reads were generated by applying a base-calling pipeline [Sequencing Control Software (SCS), Illumina]. Raw paired-end reads were aligned to the Chinese long inbred line 9930 reference genome (Huang et al. 2009) using the program Bowtie 2 (Langmead and Salzberg 2012).
Sequence analysis to detect polymorphism between gynoecious and monoecious lines
Sequence of a 50-kb region including the 30.2-kb duplication which defines the F locus on cucumber chromosome 6 of 5 gynoecious lines: TH169, WDSGD, WFEJ, WJE F11 and North American inbred line ‘Gy14’ (genome annotation is available at http://www.phytozome.net/cucumber.php#A) and 5 monoecious lines: TH118FLM, WMEJ, WNE F8, Chinese long inbred line ‘9930’ (Huang et al. 2009) and North-European Borszczagowski cultivar ‘B10’ (Woycicki et al. 2011) were analyzed using Geneious Pro7.1.3 (Biomatters, Auckland, New Zealand). Firstly, sequences of five accessions of each group were aligned using the multiple sequence alignment program, MAFFT, and then two consensus sequences derived from each sequence group were analyzed to detect the sequence polymorphisms between gynoecious and monoecious lines to develop the molecular marker linked to F locus.
DNA extraction and genotyping
Genomic DNA from individual plant was isolated from young leaves and extracted according to Dellaporta et al. (1983) with minor modifications for F 2 populations, and using DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) for inbred lines, total DNA was resuspended in 100 µL of sterile distilled water and concentration was checked after electrophoresis in 1 % agarose gels with 1× TAE buffer and stained with SYBR green.
Total twelve molecular markers were used in this study; all SSR markers were previously published (Fazio et al. 2002; Ren et al. 2009). Additional three Indel markers; Cs-Female1, Cs-Female4 and Cs-Female7 (Table 1) were designed based on the sequence polymorphisms between gynoecious inbred line, Gy14 (http://www.phytozome.net/cucumber.php#A) and monoecious Chinese long inbred line ‘9930’ from the cucumber genomics database (http://www.icugi.org). A pair of specific primers for F locus, Cs-BCAT-F/Cs-BCAT-R, which was designed based on an insertion/deletion (In/Del) polymorphism between monoecious and gynoecious lines on the analyzed 50-kb genomic region. Furthermore, a dominant marker Cs-ACS1G-pro, which is spanning the breakpoint of duplication between Cs-ACS1 and part of Cs-BCAT genes, previously developed by Wu et al. (2012) was used to detect the presence or absence of 30.2-kb duplication.
PCR reactions were in a final volume of 20 µL with 5 pmol of each primer, 20 ng of DNA and 2× Premix solution Genet Bio [Prime Taq DNA Polymerase 1 unit/10 μL, Tris–HCl (pH 9.0), PCR enhancer, (NH4)2SO4, 4 mM MgCl2, enzyme stabilizer, sediment, loading dye, 2.0 mM dNTPs mixture]. Amplification was carried out in a BIORAD T100™ Thermocycler (Hercules, CA, USA) as follows: an initial cycle at 94 °C for 3 min, followed by 35 cycles at 94 °C for 30 s, 50–57 °C (for different markers) for 30 s and 72 °C for 1 min, and a final cycle at 72 °C for 5 min. Amplified fragments were visually analyzed after electrophoresis in 2 % Agarose gel with 0.5× TBE buffer and stained with SYBR green. The PCR reactions for specific primers, Cs-BCAT-F/Cs-BCAT-R, were as above, except the annealing temperature is 63 °C. The PCR reactions for Cs-ACS1G-pro were also as above, except the annealing temperature is 57 °C with 2-min extension time for each cycle.
Linkage mapping
To perform linkage analysis of the new co-dominant markers in relation to the F locus, we analyzed the F 2 segregating population (WJE F11/WNE F8) using six SSR markers and four Indel markers on cucumber chromosome 6 (Table 1). All F 2 plants were genotyped using total ten markers and evaluated for sexual phenotype. Linkage relationships between the F locus and the molecular markers were calculated using MAPMAKER 3.0 program (Lander et al. 1987) with the Kosambi’s mapping function (Kosambi 1944). Linkage map was determined with a minimum logarithm of the odds (LOD) ratio of 3.0 and a maximum recombination fraction of 0.5.
For linkage mapping in another F 2 segregating population (WJE F10/WNE F8), 60 F 2 plants were genotyped using two SSR and four Indel markers (Table 1). Recombination values between the F locus and the molecular markers were estimated using the maximum–likelihood equation (Allard 1956) and transformed into genetic map distances (cM) using Kosambi’s mapping function (Kosambi 1944).
Results
Evaluation of sex expression
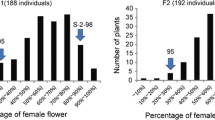
The female parents of two F 2 populations (WJE F10/WNE F8 and WJE F11/WNE F8), ‘WJE F10’ and ‘WJE F11’ showed 100 % pistillate flowers and the common male parent ‘WNE F8’ showed 41 % pistillate flowers, while their F 1 hybrids showed 100 % pistillate flowers similar to female parents. The F 2 population (WJE F11/WNE F8) consisting of 160 individuals, segregated into 119 gynoecious and 41 monoecious plants, which fits well to a 3:1 ratio (Table S2). Another F 2 population (WJE F10/WNE F8) consisting of 60 individuals, segregated into 36 gynoecious and 24 monoecious plants, which does not fit the theoretical 3:1 ratio (Table S2). On the other hand, 55 diverse inbred lines were classified into 17 gynoecious, 30 monoecious, 2 subgynoecious and the remaining six lines (SJ15, SJ41, SJ43, SJ45, SJ51 and SJ55) exhibited a less stable gynoecious phenotype because they showed subgynoecious phenotype in some years, so we designated them as subgynoecious to gynoecious phenotype, according to 5 years phenotype data (Table S1).
Sequence analysis of 50-kb genomic region of gynoecious and monoecious lines
Since F locus encodes an additional copy of Cs-ACS1 gene, Cs-ACS1G and the 30.2-kb duplication involving Cs-ACS1G defines the F locus, we analyzed the 50-kb genomic sequence including 30.2-kb duplication from five accessions of each of the two different sex types: gynoecious and monoecious. The sequence analysis showed a total of 71 In/Del polymorphisms ranging from 1 to 76 bp and 109 single nucleotide polymorphisms (SNPs) (Fig. 1; Table S3a). Among 25 insertions and 46 deletions in gynoecious lines compared with monoecious lines, seven insertions and ten deletions were conserved in all five gynoecious lines; two deletions were located in the intron of Cs-BCAT gene, one insertion and one deletion were located in the 3′-UTR of Cs-BCAT gene and the remaining six insertions and seven deletions were located in the noncoding regions (Fig. 1; Table S3a). Among 109 SNPs, 50 SNPs were conserved in all five gynoecious lines: 1 SNP and 3 SNPs were located in the 6th exon and 3′-UTR of Csa6M496980 gene, respectively, 5 SNPs, 2 SNPs and 1 SNP were located in the intron, 3′-UTR and 5′-UTR of Cs-BCAT gene, respectively, and the remaining 38 SNPs were located in the noncoding regions (Fig. 1; Table S3a). Among them, the In/Del of 3′-UTR of Cs-BCAT gene was the significant conserved (the most lengthy) polymorphism between gynoecious and monoecious lines (Table S3b).
Genomic DNA structure of monoecious and gynoecious lines of the analyzed 50-kb region on chromosome 6. Black boxes represent the genes including introns and exons. Bent arrows indicate the directions of genes. White arrowheads above and below the line indicate the conserved insertion and deletion of nucleotides, respectively, and vertical lines show conserved nucleotide differences between monoecious and gynoecious lines. Black arrowheads represent the locations and directions of the F locus-specific primers Cs-BCAT-F and Cs-BCAT-R
The sequences of four gynoecious and three monoecious lines reported in this paper were deposited in the Genbank/NCBI database under Accession Nos. KT885085, KT885086, KT885087, KT885088, KT885089, KT885090 and KT885091.
Development of an F locus-specific co-dominant marker
A pair of specific primers was designed based on a 56-bp In/Del, which located on the 3′-UTR region of BCAT gene, conserved in all five monoecious and gynoecious lines (Fig. 2), to examine the polymorphism of the parents, their F 1 hybrids and F 2 populations as well as to construct the linkage map conferring the F locus. The polymorphism of the primer pairs Cs-BCAT-F/Cs-BCAT-R was analyzed using the parents and F 1 hybrids. The PCR fragments exhibited a polymorphism between parents, with an approximately 216-bp band in the monoecious but an approximately 160-bp band in the two gynoecious lines while F 1 plants amplified both bands (Fig. 3a). This result showed that the 160- and 216-bp bands indicated homozygous-dominant genotype (FF) and homozygous recessive genotype (ff), respectively, while both bands were given by heterozygous (Ff).
Multiple alignment of the nucleotide sequences of 3′-UTR region of Cs-BCAT gene in 5 gynoecious lines (WFEJ, WJE F11, American inbred line ‘Gy14’, WDSGD and TH169) and 5 monoecious lines (Chinese long inbred line ‘9930’, TH118FLM, WMEJ, WNE F8 and North-European Borszczagowski cultivar ‘B10’). Black background indicates nucleotide sequences conserved among the accessions. Gray background indicates 56-bp deletion in gynoecious lines. Black arrows show the locations and directions of the specific primers Cs-BCAT-F and Cs-BCAT-R
The banding pattern was conserved when the two F 2 populations were analyzed using the specific primer pair. In the F 2 population (WJE F11/WNE F8), all the 41 monoecious individuals showed the 216-bp band, while among 119 gynoecious individuals, 40 of them had only one 160-bp band, but 79 individuals had two bands from 216 to 160 bp (partially shown in Fig. 3a; Table S2). In the F 2 population (WJE F10/WNE F8), all the 24 monoecious individuals showed the 216-bp band, while among 36 gynoecious individuals, 12 plants had only one 160-bp band and 24 plants showed two bands (Table S2). These results indicated that the PCR fragments of 160 and 216 bp amplified with Cs-BCAT-F/Cs-BCAT-R co-segregated with the F/f gene, behaving as a co-dominant molecular marker with two alleles: F (gynoecious) and f (monoecious).
Construction of linkage map
To verify the F locus specificity of the co-dominant marker (Cs-BCAT), linkage analysis was performed in F 2 segregating population (WJE F11/WNE F8) using six SSR markers and four Indel markers; Cs-BCAT, Cs-female1, Cs-female4 and Cs-female7, on the chromosome 6 (Table 1; Fig. 4a). The segregation ratio of gynoecious to monoecious plants (119:41) in F 2 population fits the theoretical 3:1 ratio (χ 2 = 0.033, P = 0.855) expected for dominant inheritance of a gene which promotes the femaleness in cucumber. All of the gynoecious plants were homozygous (GG) and heterozygous gynoecious (GM), whereas all monoecious plants were homozygous monoecious (MM) based on the genotype at the marker Cs-BCAT (Table S4). The observed segregation ratio of genotypes fits the theoretical 1:2:1 ratio (χ 2 = 0.038, P = 0.981) at Cs-BCAT marker, expected for the dominant inheritance of single gene related to femaleness in cucumber (Table S4). Linkage analysis showed that a single gene for femaleness co-segregated with the markers Cs-BCAT, Cs-Female1 and Cs-Female4, and was located between SSR18956 and Cs-Female7, at distances of 0.3 and 2.3 cM, respectively (Fig. 4a). This result confirms that F locus was tightly linked to Cs-BCAT marker on the chromosome 6 and enhanced femaleness in cucumber.
In another F 2 population (WJE F10/WNE F8), two SSR markers and four new Indel markers were used for linkage analysis. The observed segregation ratio of gynoecious to monoecious plants (36:24) in this population does not fit the theoretical 3:1 ratio (χ 2 = 7.2, P = 0.0073). It might be because of small population size. All gynoecious plants were homozygous (GG) and heterozygous (GM) gynoecious, whereas all monoecious plants were homozygous monoecious (MM) at the marker Cs-BCAT (Table S5). The observed segregation ratio of genotypes is slightly significant from the theoretical 1:2:1 ratio (χ 2 = 7.2, P = 0.03) at Cs-BCAT marker (Table S5) because small population size would affect the segregation ratio. Linkage analysis indicated that F locus co-segregated with Cs-BCAT and was located between Cs-Female1 and Cs-Female7, at distances of 0.8 and 2.5 cM, respectively (Fig. 4b). In this population, Cs-Female1 and Cs-Female4 were located at distances of 0.8 and 1.6 cM, respectively, from the F locus. Two previously reported SSR markers linked to the gynoecious locus; SSR18956 and CSWCT25 (Zhou et al. 2013) were located at distances of 2.5 and 5.9 cM, respectively, from the F locus.
Screening of diverse cucumber inbred lines
To further confirm the consistency of F locus-specific co-dominant marker (Cs-BCAT), we tested 55 inbred lines with different sexual phenotypes from different origins. 28 accessions had 160-bp band and among them, 17 accessions showed gynoecious, six accessions showed subgynoecious to gynoecious; but five accessions showed monoecious phenotype according to 5-year phenotype data (partially shown in Fig. 3b; Table S1). In contrast, 27 accessions had 216-bp band, and among them, 25 accessions showed monoecious and two accessions showed subgynoecious phenotypes (Table S1). Therefore, Cs-BCAT-based marker can explain ~90 % of accessions; however, ~10 % show discrepancy in which 5 accessions with homozygous gynoecious genotype at the Cs-BCAT marker showed monoecious phenotype. In two new co-dominant markers Cs-Female1 and Cs-Female4, 9 accessions and 10 accessions, respectively, showed inconsistency between marker genotypes and sexual phenotypes (Table S1).
Detection of presence or absence of 30.2-kb duplication and comparison with F locus-specific co-dominant marker
Since the F locus-specific co-dominant marker was located in the 30.2-kb duplication, we examined the presence or absence of 30.2-kb duplication in two F 2 segregating populations and additional 55 cucumber accessions using a dominant marker, Cs-ACS1G-pro (Wu et al. 2012). In both F 2 populations, all F 2 individuals showed the consistency of presence or absence of the duplication with sexual phenotypes (partially shown in Fig. S1; Table S6). Moreover, the co-dominant marker, Cs-BCAT, absolutely co-segregated with Cs-ACS1G-pro which detects the presence or absence of duplication; the presence of duplication corresponded to homozygous and heterozygous gynoecious, whereas the absence of duplication corresponded to homozygous monoecious at Cs-BCAT marker (Table S7). In additional 55 diverse accessions, the presence or absence of the duplication perfectly associated with gynoecious and subgynoecious to gynoecious, or monoecious phenotypes; however, two subgynoecious lines (SJ22 and SJ23) lacked the duplication (Table S1). Among 55 cucumber inbred lines, the Cs-BCAT marker co-segregated with Cs-ACS1G-pro in 50 accessions; however, five accessions showed homozygous gynoecious genotype at Cs-BCAT marker, but no duplication occurred. These results show that the F locus-specific Cs-BCAT marker perfectly correlated with the dominant marker, Cs-ACS1G-pro, in two F 2 segregating populations, whereas some diverse inbred lines show discrepancy between co-dominant and dominant marker genotypes. Although Cs-ACS1G-pro marker co-segregated with the sexual phenotype in both F 2 populations and diverse inbred lines, the applicability of this marker is still limited because it cannot distinguish homozygous and heterozygous gynoecious in segregating populations. The Cs-BCAT marker can distinguish perfectly homozygous and heterozygous gynoecious in the F 2 segregating populations, and it could be used for marker-assisted selection in cucumber breeding program.
Discussion
One of the primary advantages of gynoecy is to facilitate cucumber hybrid seed production by ensuring all harvested seeds are from outcrossing. Since the F locus which enhances femaleness is a partial dominant gene (Perl-Treves and Rajagopalan 2006), heterozygous (Ff) plants can exhibit a less stable gynoecious phenotype compared with homozygous (FF) plants in field conditions. Nowadays, many studies had well characterized the F locus. In addition, the dominant markers and SSR markers linked to F locus have been previously reported. However, the applicability of those makers is still limited for marker-assisted selection in cucumber breeding program because dominant markers cannot distinguish homozygous and heterozygous gynoecious lines and SSR markers were located at several genetic distances from the F locus.
Here, we reported an F locus-specific co-dominant marker which can completely distinguish homozygous and heterozygous gynoecious individuals. The marker genotypes are absolutely corresponded with the sexual phenotypes in two different F 2 populations. This marker has complete linkage to the F/f locus alleles in the present study. Among the tested cucumber accessions, some accessions showed subgynoecious phenotype whether the homozygous gynoecious (FF) or homozygous monoecious (ff) at F locus-specific co-dominant marker. Therefore, the co-dominant marker could not explain well the subgynoecious phenotype in cucumber. The genetic mechanism of subgynoecium in cucumber had rarely been described. Until now, two subgynoecious genes (one recessive gene and one dominant gene) which enhance the intensity of femaleness were reported and inherited independently with F and M genes (Chen et al. 2011). Recently, a major quantitative trait locus conferring subgynoecy in cucumber has been identified and delimited to a genomic region of 799-kb on chromosome 3 (Bu et al. 2015). It could be speculated that subgynoecious trait inherits independently with F and M genes. Our data also support the hypothesis that subgynoecious trait inherits independently with F gene because subgynoecious occurred nevertheless the genotype at F locus, such as FF or ff, in the present study.
When we examined the correlation of the F locus-specific co-dominant marker, Cs-BCAT and the dominant marker, Cs-ACS1G-pro, the co-dominant marker absolutely co-segregated with the dominant marker except the dominant marker cannot distinguish homozygous and heterozygous gynoecious in the two F 2 populations. When using diverse cucumber accessions, the presence or absence of duplication corresponded to homozygous gynoecious or homozygous monoecious at the co-dominant marker, respectively, in most cases with some exceptions in which some accessions with homozygous gynoecious genotype at the co-dominant marker showed monoecious phenotype. It could be speculated that the 56-bp deletion in these accessions might occur before 30.2-kb duplication during the evolutionary process, and it needs further investigation to explain this divergence. For such accessions, the F locus-specific marker combination (Cs-BCAT and Cs-ACS1G-pro) would be better applicable for selection of cucumber breeding lines. We further verified the F locus specificity of the co-dominant marker by linkage analysis using two F 2 segregating populations. The F locus-specific co-dominant marker showed complete linkage to F locus. Taken all together, the F locus-specific co-dominant marker could enable us to distinguish between homozygous and heterozygous gynoecious individuals and for faster and more efficient selection of cucumber breeding lines in contrast with the previously reported dominant markers and SSR markers for F locus because no recombination events occurred between the F locus-specific co-dominant marker and the F locus. It is also suitable for high-throughput screening or marker-assisted selection in cucumber breeding program because of its low cost and convenient PCR operation.
So far, no functional evidence of F gene in cucumber was revealed even though several studies have been done for the F locus in cucumber. Because 30.2-kb duplicated region including the additional copy of Cs-ACS1 (Cs-ACS1G) is defined as the F locus (Zhang et al. 2015) and we found the F locus-specific co-dominant marker is located on the 3′-UTR region of Cs-BCAT on chromosome 6 in this study, an important question is whether Cs-BCAT involves in determination of cucumber sex types. In Arabidopsis, BCATs encoded by at least six genes (BCAT1 to BCAT6) act as aminotransferase proteins in Ile, Leu and Val metabolic pathways (Diebold et al. 2002). However, non-rescued phenotype of yeast mutant lacking endogenous BCAT activity by BCAT4 (Less and Galili 2008; Schuster and Binder 2005) and significant amino acid similarity of Arabidopsis BCAT with yeast Bat1p or Bat2p acting in 2-keto-4-methyl-thiobutyrate (KMTB) transamination of methionine (Met) salvage pathway (Pommerrenig et al. 2011) suggests that BCAT may function in recycling of Met during ethylene biosynthesis in plants. This possibility is supported by the observations that the expression levels of Arabidopsis BCAT homologs in banana and melon were changed during fruit ripening or with ethylene treatment, respectively (Yan 2012; Yang et al. 2011). However, the function of Cs-BCAT homologs has not yet to be determined in cucumber, even though cucumber genome has at least three Arabidopsis BCAT homologs. Thus, further investigation is required to elucidate the role of Cs-BCAT in ethylene biosynthesis pathway in cucumber. Also, further investigation on the role of the 56-bp In/Del variation at 3′-UTR region of Cs-BCAT would provide a better understanding on determination of reproductive morphology in cucumber.
References
Allard RW (1956) Formulas and tables to facilitate the calculation of recombination values in heredity. Hilgardia 24:235–278
Bu F, Chen H, Shi Q, Zhou Q, Gao D, Zhang Z, Huang S (2015) A major quantitative trait locus conferring subgynoecy in cucumber. Theor Appl Genet. doi:10.1007/s00122-015-2612-z
Chen H, Tian Y, Lu X, Liu X (2011) The inheritance of two novel subgynoecious genes in cucumber (Cucumis sativus L.). Sci Hortic (Amsterdam) 127:464–467
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: Version II. Plant Mol Biol Rep 1:19–21
Diebold R, Schuster J, Daschner K, Binder S (2002) The branched-chain amino acid transaminase gene family in Arabidopsis encodes plastid and mitochondrial proteins. Plant Physiol 129:540–550
Fazio G, Staub JE, Chung SM (2002) Development and characterization of PCR markers in cucumber. J Am Soc Hortic Sci 127(4):545–557
Huang S, Li R, Zhang Z, Li L, Gu X, Fan W, Lucas WJ, Wang X, Xie B, Ni P (2009) The genome of the cucumber, Cucumis sativus L. Nat Genet 41:1275–1281
Knopf RR, Trebitsh T (2006) The female-specific CsACS1G gene of cucumber. A case of gene duplication and recombination between the non-sex-specific 1-aminocyclopropane-1-carboxylate synthase gene and a branched chain amino acid transaminase gene. Plant Cell Physiol 47(9):1217–1228
Kosambi D (1944) The estimation of map distance from recombination values. Ann Eugen 12:172–175
Kubicki B (1969) Investigation on sex determination in cucumber (Cucumis sativus L.). Genet Pol 10:69–86
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:178–181
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Less H, Galili G (2008) Principal transcriptional programs regulating plant amino acid metabolism in response to abiotic stresses. Plant Physiol 147:316–330
Malepszy S, Niemirowicz-Szczytt K (1991) Sex determination in cucumber (Cucumis sativus) as a model system for molecular biology. Plant Sci 80:39–47
Mibus H, Tatlioglu T (2004) Molecular characterization and isolation of the F/f gene for femaleness in cucumber (Cucumis sativus L.). Theor Appl Genet 109:1669–1676
Perl-Treves R (1999) In sex determination in plants. In: Ainsworth C (ed) Male to female conversion along the cucumber shoot: approaches to studying sex genes and floral development in Cucumis sativus. Blackwell, London, pp 189–216
Perl-Treves R, Rajagopalan PA (2006) Annual plant reviews volume 20: flowering and its manipulation. In: Ainsworth C (ed) Close, yet separate: patterns of male and female floral development in monoecious species. Blackwell, London, pp 117–146. doi:10.1002/9780470988602.ch6
Pommerrenig B, Feussner K, Zierer W, Rabinovych V, Klebl F, Feussner I, Sauer N (2011) Phloem-specific expression of Yang cycle genes and identification of novel Yang cycle enzymes in Plantago and Arabidopsis. Plant Cell 23:1904–1919
Ren Y, Zhang Z, Liu J, Staub JE, Han Y, Cheng Z (2009) An integrated genetic and cytogenetic map of the cucumber genome. PLoS One 4(6):e5795. doi:10.1371/journal.pone.0005795
Schuster J, Binder S (2005) The mitochondrial branched-chain aminotransferase (AtBCAT-1) is capable to initiate degradation of leucine, isoleucine and valine in almost all tissues in Arabidopsis thaliana. Plant Mol Biol 57:241–254
Shifriss O (1961) Sex control in cucumbers. J Hered 51:5–12
Trebitsh T, Staub JE, O’Neill SD (1997) Identification of a 1-aminocyclopropane-1-carboxylic acid synthase gene linked to the Female (F) locus that enhances female sex expression in cucumber. Plant Physiol 113:987–995
Woycicki R, Witkowicz J, Gawronski P, Dabrowska J, Lomsadze A (2011) The genome sequence of the North-European cucumber (Cucumis sativus L.) unravels evolutionary adaptation mechanisms in plants. PLoS One 6(7):e22728
Wu T, Qin Z, Feng Z, Zhou X, Xin M, Du Y (2012) Functional analysis of the promoter of a female specific cucumber CsACS1G gene. Plant Mol Biol Rep 30:235–241. doi:10.1007/s11105-011-0318-1
Yan L (2012) Effect and regulation of ethylene in the biosynthesis of aroma volatiles in oriental sweet melon (Cucumis melo var. Maruwa Makino). Shenyang Agricultural University, China
Yang XT, Song J, Fillmore S, Pang XQ, Zhang ZQ (2011) Effect of high temperature on color, chlorophyll fluorescence and volatile biosynthesis in green-ripe banana fruit. Postharvest Biol Technol 62:246–257
Zhang Z, Mao L, Chen H et al (2015) Genome-wide mapping of structural variations reveals a copy number variant that determines reproductive morphology in cucumber. Plant Cell. doi:10.1105/tpc.114.135848
Zhou S, Zhang P, Zhu Y, Chen X, Chen L (2013) Identification of SSR marker linked to gynoecious loci in cucumber (Cucumis sativus L.). J Zhejiang Univ Agric Life Sci 39(3):291–298. doi:10.3785/j.issn.1008-9209.2012.11.141
Acknowledgments
This work was supported by Grants from Bio-industry Technology Development Program (111057-5) of iPET (Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Win, K.T., Zhang, C., Song, K. et al. Development and characterization of a co-dominant molecular marker via sequence analysis of a genomic region containing the Female (F) locus in cucumber (Cucumis sativus L.). Mol Breeding 35, 229 (2015). https://doi.org/10.1007/s11032-015-0424-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-015-0424-0