Abstract
NAC proteins constitute a family of plant-specific transcription factors that are involved in many plant cellular processes including responses to abiotic stress. In this study, a cDNA clone encoding the HvSNAC1 transcription factor was isolated from drought-stressed barley using a bioinformatics approach based on amino acid sequence data of the stress-related SNAC1 protein from rice. Phylogenetic analysis of the deduced amino acid sequence of HvSNAC1 showed that this protein belongs to the stress clade of NAC proteins that include SNAC1 and TaNAC2. Expression analysis indicated that the HvSNAC1 gene is strongly induced by different abiotic stresses including drought. Overexpression of HvSNAC1 in barley under the control of a constitutive promoter produced plants that grew normally under well-watered conditions when compared with wild-type plants. Transgenic barley plants overexpressing HvSNAC1 showed higher drought tolerance at different growth stages when compared with wild-type plants. In addition, the constitutive overexpression of HvSNAC1 resulted in improved water status, photosynthetic activity and reduced water loss rate when compared with wild-type plants under drought conditions. Furthermore, the transgenic plants also showed significantly improved productivity, as reflected by the increase in biological yield over the wild-type plants under severe field drought conditions. In conclusion, the HvSNAC1 gene could be a useful tool for improving barley productivity under field drought conditions without impairment in growth under normal field conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Barley (Hordeum vulgare L.) is an important cereal plant that is mainly cultivated in marginal lands where adverse environmental conditions such as drought and high temperatures are frequent. In such areas, there is an urgent need to develop drought-tolerant plants that can withstand such adverse conditions while maintaining high productivity (Habash et al. 2009). Plant breeding for improved drought-tolerant crops has demonstrated that this complex trait is governed by multiple loci (Mir et al. 2012). This inherited genetic complexity and redundancy of tolerance mechanisms have made the identification of key regulatory genes governing drought tolerance in plants a priority.
Several studies were conducted to understand the mechanisms by which plants perceive environmental signals to activate cellular mechanisms involved in stress-tolerance responses (Bartels and Sunkars 2005). In this perspective, drought-related signals were found to activate large numbers of regulatory genes including many transcription factors and kinases (Zhu 2002). The activation of these genes results eventually in the activation of numerous stress-related genes and the synthesis of diverse metabolites that improve plant tolerance to drought. For instance, studying gene expression changes associated with abiotic stresses in different plants revealed that several transcription factors were involved in plant responses to drought (Seki et al. 2001; Bhargava and Sawant 2013). Many of these transcription factors were found to be members of different families, including AP2/EREBP, bZIP, NAC, MYB, MYC, zinc-finger and WRKY (Yamaguchi-Shinozaki and Shinozaki 2006; Nakashima et al. 2009). In many instances, the overexpression of these factors or their active forms in plants confers cold, drought and salt tolerance phenotypes (Umezawa et al. 2006; Xue et al. 2011; Mao et al. 2012).
One of these transcription factors families is the NAC gene family, which comprises more than 100 plant-specific transcription factors that are involved in various developmental and stress responses (Olsen et al. 2005; Puranik et al. 2012). NAC proteins are characterized by their highly conserved DNA binding domain in the N-terminal region, which was originally identified from consensus sequences of petunia NAM and Arabidopsis ATAF1, ATAF2 and CUC2 genes (Aida et al. 1997). NAC proteins play important roles in abiotic and biotic responses including viral resistance (Ren et al. 2000), bacterial infection and wounding (Collinge and Boller 2001; Mysore et al. 2002; Hegedus et al. 2003), and salt stress and drought tolerance (Fang et al. 2008; Nakashima et al. 2012). For instance, a stress-inducible rice NAC gene (SNAC1) was found to play a major role in stress tolerance and its overexpression resulted in drought tolerance of field-growing rice plants at anthesis (Hu et al. 2006). The overexpression of another stress-inducible rice NAC gene (SNAC2) improved drought tolerance at seedling stage; however, testing the SNAC2 overexpression rice lines in the field under drought conditions did not have any significant effect on yield (Hu et al. 2008). The constitutive overexpression of another NAC gene (OsNAC6) in rice resulted in improved drought tolerance at the seedling stage; however, pleiotropic effects of vegetative growth were observed (Nakashima et al. 2007). On the other hand, overexpression of OsNAC6 using stress-inducible promoter resulted in improved stress tolerance in transgenic rice plants while minimizing negative pleiotropic effects on plant growth. The root-specific expression of OsNAC10 improved drought tolerance and yield in field-grown rice plants (Jeong et al. 2010). In barley, HvNAC6 was found to enhance resistance against pathogen penetration (Jensen et al. 2007), while rendering tolerance against drought stress (Lu et al. 2007). The characterization of 48 barley NAC genes suggested that they are functionally conserved with other related NAC genes from other plant species (Christiansen et al. 2011). Such reports indicate that NAC genes play a pivotal role in mediating responses to different environmental stimuli and confirm their potential utilization to enhance stress tolerance including drought. In many instances, the overexpression of the NAC transcription factors in different plant species resulted in improved stress tolerance toward freezing and dehydration tolerance (Fang et al. 2008). Such data suggest that stress-responsive NAC transcription factor overexpression has the potential to enhance drought tolerance in cultivated plants including barley. Accordingly, it seems feasible to utilize genetic information of previously characterized key regulatory stress-responsive NAC genes from model plants to clone candidate genes in barley. The isolation of NAC candidate genes in barley can be utilized to generate transgenic plants with enhanced tolerance to drought stress. However, it is still not clear what would be the influence of the overexpression of these genes on the physiological processes under water deficit conditions.
To answer these questions, the current study was initiated in order to isolate and characterize a stress-related drought-tolerance NAC gene from barley. The HvSNAC1 gene was cloned and found to be highly related to stress-responsive NAC genes from grasses. HvSNAC1 was induced in response to drought conditions, indicating a potential role in mediating tolerance to dehydration. Transgenic plants overexpressing HvSNAC1 showed normal growth and reduced water loss rate under drought conditions. In addition, HvSNAC1 overexpression lines improved yield of barley under drought field conditions.
Materials and methods
Cloning of a stress-related NAC gene in barley
Based on the DNA sequence of the previously described SNAC1 gene in rice (GenBank accession number: DQ394702), a TBLASTN search with the amino acid sequence of SNAC1 protein was performed against two nucleotide sequences databases (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi and http://pgrc.ipk-gatersleben.de/est/). Using this approach, several expressed sequence tags (ESTs) and mRNA sequences were obtained and were found to encode a putative stress-related NAC gene in barley. Using the obtained DNA sequence of Genbank accession number JF796130, a gene-specific primer pair of 5′-ATGGGGATGCCGGCGGCGAG-3′ (HvSNACfwd) and 5′-TTAGAACGGGGCTGGCATGC-3′ (HvSNACrev) was designed.
To isolate the full-length cDNA of the corresponding stress-related NAC gene, 2-week-old barley cv. Golden Promise seedlings were subjected to drought treatment by withholding water for 7 days. Whole leaf tissue was harvested from stressed plants and flash-frozen in liquid nitrogen until total RNA extraction. Total RNA was isolated from stressed tissue using SV Total RNA Isolation System Kit (Promega, Madison, WI, USA) following the manufacturer’s instructions. The isolated RNA was used to synthesize first-strand cDNA library using the SuperScript® First-Strand Synthesis System (Invitrogen, USA) and oligo T(18) primer following the manufacturer’s instructions. The full-length cDNA of stress-related barley NAC was amplified using PCR in a 25 μL reaction mixture containing 5 μL of cDNA as a template, 2.5 μL of dNTPs (100 μM), 5 μL of 5× PCR buffer, 0.5 μM of each primer and 0.25 μL of 5 U/μL GoTaq DNA polymerase (Promega). The PCR conditions were 94 °C for 5 min, followed by 40 cycles of 94 °C for 30 min, 55 °C for 1 min and 72 °C for 1 min, and a final 10 min extension at 72 °C. The amplified PCR products were separated in 1 % agarose gel stained with ethidium bromide. Positive PCR products were extracted from agarose gel using Wizard® SV Gel and PCR Clean-Up System (Promega) and cloned into pGEM®-T Easy Vector System (Promega) following the manufacturer’s instructions. The pGEM-T Easy Vector System allows the selection of positive recombinant plasmids based on the insertional inactivation of the α-peptide of the enzyme β-galactosidase. Positive recombinant plasmids that contained the full-length HvSNAC1 cDNA were fully sequenced using the M13 reverse and forward sequencing primers in a ABI 3730xl machine (Macrogen, Seoul, Korea).
Bioinformatics analysis
Sequence analysis and annotation predication was performed using the Gramene database (Youens-Clark et al. 2010) and MIPS barley genome database (http://mips.helmholtz-muenchen.de/plant/barley/index.jsp). For promoter analysis, PlantCARE (Lescot et al. 2002) and PLACE/Signal Scan (Higo et al. 1999) databases were used to mine for stress-related cis regulatory elements. In silico gene expression analysis was based on microarray expression data from various datasets using GeneVestigator software (https://www.genevestigator.com/gv/; Zimmermann et al. 2008). For multiple sequence alignments analysis, ClustalX software (version 2.0; Thompson et al. 1997) was used. A phylogenetic analysis was carried out using MEGA5 (Tamura et al. 2011). The protein sequence of the barley stress-induced NAC, together with selected NAC protein sequences from wheat and rice retrieved from GenBank, were aligned using the ClustalW algorithm in MEGA5. The alignment was used to calculate distance matrices for neighbor-joining analyses with the Kimura two-parameter model. Bootstrap analysis with 1,000 replicates was performed to test the robustness of the internal branches.
Semi-quantitative and quantitative real-time RT-PCR analysis
Semi-quantitative RT-PCR analysis was performed using first-strand cDNA prepared from barley plants using gene-specific and internal reference gene primers. For this purpose, a HvSNAC1 gene-specific primer pair 5′-GGATTCCGCTTCCACCCCAC-3′ and 5′-ATCATGGTCTCCCCTTCCCC-3′ was designed for expression analysis. A fragment of barley HvActin gene (accession number AY145451) was amplified with the primer pair 5′-CTCCTTCACAACCTCAGCTG-3′ and 5′-CAAGGTCGTCGCTCCACCTG-3′ as an internal control for the relative amount of RNA. For semi-quantitative RT-PCR, the PCR conditions were as described above and modified as follows: denaturation for 5 min at 95 °C, followed by 30–35 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s and elongation at 72 °C for 60 s, and a final elongation at 72 °C for 10 min.
For quantitative real-time PCR analysis, total RNA was isolated from leaf samples taken from Golden Promise seedlings drought-stressed for 3 and 5 days and non-stressed. Specific primers pairs for HvSNAC1, HvActin (used as the reference gene), HVA1, HVA22, DHN8 and HvCor14b (considered as drought-inducible genes) were designed using Primer 3 software. The amplification of the targeted genes was carried out using the GoTaq® qPCR Master Mix Kit (Promega), and real-time detection of products was performed in a Mini Opticon Real Time PCR System (Bio-Rad, Hercules, CA, USA). All cDNA samples were analyzed in triplicate, and the cDNA was derived from at least two biological replicates. Thermal cycling conditions consisted of 40 cycles of 94 °C for 30 s, 58 °C for 30 s and 72 °C for 45 s, plus a final extension at 72 °C for 5 min. The relative changes in gene expression were quantified as described in Vandesompele et al. (2002).
Plant transformation
To generate transgenic barley plants overexpressing the stress-related HvSNAC1 gene, the verified coding sequence of HvSNAC1 was inserted downstream of the maize Ubi promoter in the pBRACT211 binary plasmid. In addition, the coding sequence of HvCBF3 (accession number AF239616), a drought-responsive transcription factor conferring tolerance in overexpression transgenic barley plants (Choi et al. 2002), was introduced into the same binary plasmid to generate positive control lines. The cloned plasmids were introduced into barley cv. Golden Promise via Agrobacterium tumefaciens-mediated transformation as described previously (Bartlett et al. 2008). Positive transgenic plants were verified using PCR and antibiotic resistance screening assay. T1 plants underwent further analysis for transgene presence using PCR as described in Bartlett et al. (2008), screening for antibiotic resistance plants as described in Wang and Waterhouse (1997), and gene expression levels using quantitative real-time PCR analysis using first-strand cDNA library prepared from total RNA isolated transgenic plants as described above. Two T1 plants showing high levels of HvSNAC1 expression were selected to obtain homozygous T2 plants by screening for antibiotic resistance plants as described in Wang and Waterhouse (1997). The T2 homozygous plants from both lines were further selected to produce T3 homozygous plants that were then used for the stress experiments described below.
Greenhouse experiments
Drought tolerance at the seedling stage was evaluated in sand culture in small pots. For this purpose, 15 seeds from each positive homozygous line and non-transgenic control were planted in small pots (10 cm length and 10 cm diameter). After seedling emergence, the seedlings were fed with Hoagland solution for 2 weeks. After 2 weeks, drought condition was imposed by continuous water withholding for 10 days followed by re-watering. The wilting behavior of the treated plants and the survival rates of transgenic lines and the non-transgenic control were monitored.
For continuous drought-stress treatment on plants at flag leaf stage, seeds of both transgenic and wild-type barley plants were grown in pots. The pots were filled with a mixture of soil and perlite and one seed of each genotype was sown in the pot. The pots were then irrigated to bring initial soil moisture to field capacity. After seedling emergence, the pots were exposed to two water regimes: “non-stressed”, in which irrigation water was applied when soil moisture reached a level of 20 % water depletion of field capacity, and “stressed”, in which irrigation was applied 7 days after soil moisture reached 50 % water depletion of the field capacity. The treated plants were counted for final tiller number at the flag leaf stage.
For physiological measurements, leaf relative water content was measured under control (well-watered plants) and drought-stress conditions (described above) for transgenic and wild-type plants. For this purpose, fully expanded leaves were taken from transgenic and wild-type plants grown in the same pot at two different growth stages (onset of stem elongation and flag leaf emergence) and their fresh weights were immediately recorded after leaf excision. The excised leaves were soaked in distilled water for 6 h at room temperature in darkness and the turgid weight (TW) was recorded. Total dry weight (DW) was then recorded after drying for 24 h at 70 °C. RWC was calculated according to Barrs and Weatherley (1962): RWC (%) = [(FW − DW)/(TW − DW)] × 100. Stomatal resistance was measured in greenhouse-grown seedlings at approximately midday after starting water stress treatments using a porometer (AP4, Delta-T Devices, Cambridge, UK) on three selected leaves per barley genotype. Chlorophyll fluorescence parameters were also measured after starting water stress treatments with a pulse-modulated fluorometer (OS1-FL modulated chlorophyll fluorometer, ADC BioScientific Ltd., Hertford, UK). Fluorescence measurements were made after 20 min of acclimation to darkness under leaf clips (FL-DC; Opti-Sciences). The maximum quantum efficiency (F v/F m) of photosystem II was calculated as F v/F m = (F m − F 0)/F m, where F m and F 0 were the maximal and minimal fluorescence measured in darkness-adapted leaves, respectively (Maxwell and Johnson 2000).
Field trials
Transgenic plants and wild-type control were grown under rainfed conditions at the experimental station of the Jordan University of Science and (JUST) in Ramtha, Jordan in the 2011–2012 growing season. The JUST station represents a drought-prone area in the northern part of Jordan with an annual precipitation of 200 mm. The experimental layout was prepared according to a randomized complete block design with four replications. In each block, seeds of the tested genotypes were sown into 2-m rows at a density of 50 seeds per meter and 50-cm row spacing. The field experiment was managed following the standard agricultural practices, including fertilizer application, weed control and pesticide use against main pathogens. At the end of the growing season, plants were harvested and yield parameters such as tiller number, total biological yield, spike weight, straw weight, spike number, grains per spike, grain number and grain yield were recorded.
Results
Isolation of stress-related NAC gene in barley
To identify the stress-related NAC gene in barley, bioinformatics analysis was carried out based on the full-length sequence data of a previously described stress-related rice SNAC1 protein (SNAC1; GenBank ID: ABD52007). For this purpose, a TBLASTN search was performed against the GenBank nucleotide database (www.ncbi.nlm.nih.gov/blast) and the IPK Crop EST database (http://pgrc.ipk-gatersleben.de/est/). As a result, several EST and mRNA sequences were retrieved and found to encode a putative SNAC1 protein in barley (HvSNAC1: GenBank ID: JF796130). The deduced complete amino acid sequence of HvSNAC1 has identity of 69 % with the rice SNAC1 protein and 97 % with TaNAC2 (AAU08786). Using a gene-specific primer pair designed from the JF796130 nucleotide sequence, a 993-bp fragment containing the coding sequence of the HvSNAC gene was obtained from drought-stressed barley tissues.
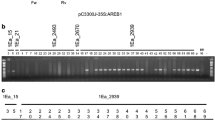
Mining the sequence data of the released barley genome in the MIPS barley genome database (The International Barley Genome Sequencing Consortium 2012: http://mips.helmholtz-muenchen.de/plant/barley/index.jsp) and Gramene database (http://www.gramene.org/Hordeum_vulgare/Info/Index) indicated that the HvSNAC1 gene was located on chromosome 5H, and it is annotated as MLOC_37104. Two splice variants were predicated, where MLOC_37104.1 encodes a truncated protein (186 amino acids) while MLOC_37104.2 encodes a functional HvSNAC1 protein (330 amino acids). In addition, several natural single nucleotide polymorphism variants were identified in the DNA sequences of the 5′ untranslated region (UTR), exons, intron and 3′ UTR (data not shown). The genomics structure of the MLOC_37104.2 variant revealed that HvSNAC1 has 1,769 bps and contained two exons and one intron, a 115-bp 5′ UTR and a 213-bp 3′ UTR. The protein architecture of HvSNAC1 indicated the presence of a NAC domain (DNA binding domain) in the N-terminal region (19–172 amino acid residues) that could be further divided into five highly conserved sub-domains (Fig. 1a). Phylogenetic analysis grouped the isolated HvSNAC1 protein with SNAC1 and TaNAC2 proteins in the stress NAC proteins clade (Fig. 1b).
a Multiple sequence alignment analysis of HvSNAC1 (AEG21060) and selected NAC proteins from wheat (AAU08786 (TaNAC2) and ADE34586) and rice (ABD52007 (SNAC1) and ACX71077). The five subdomains (A–E) of the NAC domain are underlined. b Phylogenetic analysis of selected NAC protein sequences from barley, rice and wheat. The position of HvSNAC1 protein (AEG21060) in the tree is indicated by the dashed box
HvSNAC1 is expressed in response to drought conditions
To analyze the expression patterns of HvSNAC1 under different conditions, in silico expression analysis using the gene-expression search engine, GeneVestigator (https://www.genevestigator.com/gv/user/gvLogin.jsp), was performed. As shown in Fig. 2a, the in silico gene expression in barley plants showed up-regulation in response to different abiotic stresses including drought. In addition, gene expression induction was observed in barley challenged with Fusarium graminearum. Interestingly, a significant down-regulation of HvSNAC1 expression was observed in barley plants treated with gibberellin, aphid and Blumeria graminis, the fungal agent that causes powdery mildew in cereals.
a In silico expression analysis of HvSNAC1 by GeneVestigator. b Distribution of major known stress-related cis-elements in HvSNAC1 promoter region. c Expression analysis of HvSNAC1, HVA1, HVA22, HvCor14b and DHN8 genes in responses to drought treatment after 3 and 5 days compared with well-watered samples (0 day). Values are mean ± SD of three technical replicates
In order to find cis-acting elements involved in abiotic responses, the HvSNAC1 promoter region [2,000 bp upstream of the 5′ UTR obtained from morex_contig_2547787 based on the draft barley genome sequence (http://mips.helmholtz-muenchen.de/plant/barley/index.jsp)] was mined using two plant cis-element databases, PlantCARE (Lescot et al. 2002) and PLACE (Higo et al. 1999). Using this strategy, several stress-responsive elements were identified in the HvSNAC1 promoter region, including abscisic acid responsive element (ABRE), dehydration responsive element (DRE), low temperature responsive (LTR), myeloblastosis recognition site (MYBR) and myelocytomatosis recognition site (MYCR) (Fig. 2b).
To further investigate the role of the HvSNAC1 gene in response to drought stress, expression analysis of HvSNAC1 and selected drought-inducible genes was performed in Golden Promise seedlings exposed to drought conditions for 3 and 5 days. These selected genes included HVA1, HVA22, DHN8 and HvCor14b, known for their induction under drought in barley (Morran et al. 2011). Under drought conditions, the expression of HvSNAC1 was induced after 3 days and showed higher levels of induction after 5 days of treatment when compared with wild-type plants under normal conditions (Fig. 2c). The expression patterns of HvSNAC1 were similar to the drought-inducible genes, where increased levels were observed after 3 days of drought treatment.
Overexpression of HvSNAC1 in barley enhanced drought tolerance
To test whether HvSNAC1 can improve tolerance to drought stress in barley plants, an overexpression construct with HvSNAC1 CDS under the control of maize Ubiquitin promoter was introduced into barley cv. Golden Promise. Twenty-six independent transgenic lines were obtained based on hygromycin selection, PCR analysis and expression analysis of HvSNAC1 (data not shown). The majority of the transgenic lines grew normally with no stunting when compared with non-transformed plants (Supplement Fig. 1a). The T3 generations of two homozygous lines overexpressing HvSNAC1 (lines OE#3 and OE#11) (Supplement Fig. 1b) were selected and further used for drought tolerance assay and physiological measurements.
To test drought tolerance in the selected transgenic lines at early growth stages, 2-week-old plants grown in small pots filled with sand were subjected to withholding of water for a period of 10 days prior to re-watering. The drought-subjected plants were monitored for growth and survival at different set points of the treatment. Wild-type plants were not able to survive under such adverse conditions and the majority of the stressed plants wilted and turned yellow after 10 days of treatment (Fig. 3a, b). However, the HvSNAC1 transgenic lines were able to survive such adverse conditions and over 90 % of the tested OE plants showed improved drought tolerance when compared with wild-type plants. In addition, measuring the RWC of stressed plants after 5 days of drought treatment revealed that the transgenic plant had higher values than wild-type plants (Fig. 3c). Furthermore, under water-deficit conditions (5 days of drought treatment) more stomata were found to be closed in transgenic lines than in wild-type (Fig. 3d). Interestingly, no differences in stomatal resistance were observed between transgenic and wild-type plants under normal conditions. Measuring the maximum yield of PSII (F v/F m) in well-watered plants showed no major differences among the wild-type and transgenic lines (Fig. 3e). After 5 days of drought stress, the F v/F m values were decreased in wild-type plants while transgenic lines showed no reduction in their values.
Growth performance of wild-type, transgenic line OE#3 and transgenic line OE#11 at stem elongation stage. a After 5 days of drought stress. b After 10 days of drought stress. c RWC in leaves of 2-week-old wild-type, OE#3 and OE#11 seedlings after 5 days of water withholding. d Stomatal resistance of 2-week-old wild-type, OE#3 and OE#11 seedlings after 5 days of water withholding. Values are the mean ± SE of 10 plants. e Maximum quantum yield of PSII (F v/F m) of 2-week-old wild-type, OE#3 and OE#11 seedlings after 5 days of water withholding
To test the effect of drought stress on mature plants at the flag leaf stage under greenhouse conditions, wild-type plants and T3 homozygous transgenic plants were subjected to two water regimes as described in “Materials and Methods”. Under well-watered conditions, the OE lines and wild-type plants showed normal vegetative growth, which was almost similar between the tested genotypes (Fig. 4a). Under prolonged drought conditions, both wild-type and transgenic lines suffered from the adverse conditions with a severe reduction in growth (Fig. 4a). However, under severe drought conditions, the transgenic lines showed a higher number of tillers than wild type plants (Fig. 4b). Measuring plant RWC of non-stressed plants indicated that the well-watered wild-type and transgenic plants had no significant difference (Fig. 4c). Under drought stress conditions, the RWC in the wild-type plant was significantly lower than in the OE lines (Fig. 4c). These results indicate that the OE transgenic lines had better water status than wild-type plants after exposing them to severe drought conditions.
a Growth performance of OE#3 and wild-type at flag leaf stage after continuous drought stress treatment. b Tiller number of wild-type, OE#3 and OE#11 plants at flag leaf stage after subjecting them to two different watering regimes. c RWC in leaves of wild-type, OE#3 and OE#11 plants at flag leaf stage after subjecting them to two different watering regimes. Values are the mean ± SE of three replicates
The performance of the OE lines and wild-type plants was evaluated under drought conditions in the field. T3 seeds of homozygous lines of OE#3 and OE#11 together with wild-type controls were sown under rainfed conditions in a field location characterized by low annual precipitation (215 mm), silty soil and prevalent drought conditions. In addition, the T3 homozygous lines of OE#3 and OE#11 were compared with two T4 homozygous transgenic lines overexpressing HvCBF3, a key transcription factor of cold and drought regulation in barley (Choi et al. 2002). The overexpression of CBF/DREB factors in wheat and barley was shown to improve tolerance under severe drought conditions (Morran et al. 2011). At the end of the growing season, yield parameters were recorded from four replicates. As shown in Fig. 5a, the OE#3 and OE#11 transgenic plants showed improved growth at the tillering stage in the field when compared with the wild-type plants. Comparing the yields of the tested genotypes showed that all of the transgenic lines (HvSNAC1 OE and HvCBF3 OE) produced significantly higher tiller number, which resulted in higher total weight, spike number, grains per spike, grain number and grain weight when compared with the wild-type plants (Fig. 5b). These findings clearly demonstrate that HvSNAC1 overexpression can improve yield under drought conditions in the field.
a Growth of wild-type and HvSNAC1 OE#3 and OE#11 transgenic lines in the field under drought conditions. b Agronomic performance of HvSNAC1 and HvCBF3 OE plants grown in the field under drought conditions. Each data point represents the percentage of the mean values of an agronomic trait from plants harvested from a single row
Discussion
In this study, a stress-related NAC gene was isolated and characterized and showed significant improved tolerance to drought stress when overexpressed in barley plants. Recently, 48 NAC genes were identified in barley and found to have conserved functions with related NAC genes from other plant species including developmental processes and responses to environmental stimuli (Christiansen et al. 2011). Phylogenetic analysis of HvSNAC1 showed a close relationship with previously characterized stress-related NAC genes such as SNAC1 from rice (Hu et al. 2006) and TaNAC2 from wheat (Mao et al. 2012). HvSNAC1 protein exhibited maximum sequence identity (97 %) to TaNAC2 (AAU08786), which was found to be involved in response to drought, salt, cold and abscisic acid and to improve stress tolerance when expressed in Arabidopsis plants (Mao et al. 2012). This similarity of HvSNAC1 to other stress-related NAC genes in grasses indicates a major role in mediating stress tolerance in barley. The HvSNAC1 is located on the lower arm of chromosome 5H and it is annotated as MLOC_37104. This region is well known to contain many drought-tolerant candidate genes in barley plant (Tondelli et al. 2006). Identification of HvSNAC1 beside other candidate genes involved in drought tolerance in this important region in the barley genome highlights the need to develop an efficient marker-assisted selection strategy to improve breeding activities related to the enhancement of barley productivity in drought-prone areas.
In silico expression analyses showed that the HvSNAC1 gene was induced by several abiotic stresses including drought, cold and heat. HvSNAC1 expression induction in response to dehydration stress was confirmed in drought-stressed barley seedlings. This is consistent with the in silico promoter analysis where several stress-responsive regulatory elements were identified in HvSNAC1 promoter region. Such induction patterns are similar to previously characterized stress-related NAC genes from other plant species (Hu et al. 2006; Mao et al. 2012), suggesting a role of HvSNAC1 in mediating drought tolerance in barley plants. Besides its role in abiotic tolerance, the isolated gene is expected to function in responses against biotic stresses. This assumption is supported by GeneVestigator data, where HvSNAC1 expression is altered by several pathogens. HvSNAC1 was induced in plants challenged with F. graminearum and repressed in plants challenged with B. graminis. Several NAC genes were found to have an important role in mediating responses against pathogens. For instance, a novel wheat NAC transcription factor was found to play a major role in mediating responses to both biotic and abiotic stresses (Xia et al. 2010). The HvNAC6 gene was found to act as a positive regulator of penetration resistance against B. graminis in barley (Jensen et al. 2007). In Arabidopsis, the overexpression of ATAF2, a stress-related NAC gene, resulted in higher susceptibility to the soil-borne fungal pathogen Fusarium oxysporum (Delessert et al. 2005). Furthermore, the HvSNAC1 homolog in rice (SNAC2) was found to play a major role in tolerance responses against biotic and abiotic stresses (Hu et al. 2008). This suggested that HvSNAC1 might play a major role in mediating plant responses to other kinds of stresses besides drought. In the future, the role of HvSNAC1 in mediating responses of barley to different pathogen infections will be studied.
Interestingly, the constitutive expression of HvSNAC1 in barley resulted in normal vegetative growth when compared with wild-type plants. In contrast to HvSNAC1, the overexpression of other drought-responsive transcription factors in other plants usually resulted in pleiotropic effects such as reduction in plant growth and delayed flowering (Morran et al. 2011). Such adverse growth retardation will be reflected in the final yield and plant performance under field conditions. Such results are in general agreement with previous studies where the expression of NAC genes in different plant species resulted in normal growth without major yield penalties (Hu et al. 2006, 2008; Mao et al. 2012; Saad et al. 2013). Such improved tolerance without major pleiotropic effects of HvSNAC1 overexpression is considered promising for crop improvement for dry areas.
The induction of HvSNAC1 in response to different abiotic stresses indicates the possibility of conferring enhanced tolerance to drought in HvSNAC1-overexpressing transgenic plants. Testing drought tolerance in transgenic lines indicates that HvSNAC1 overexpression enhanced drought tolerance at seedling and reproductive stages under greenhouse and field conditions. Under drought field conditions, the overexpression of HvSNAC1 in barley plants enhanced several yield components including tiller number, spike number, grain number, grain weight and grain per spike when compared with non-transgenic plants. Similar results were obtained with the overexpression of SNAC1 in rice, where no obvious differences between the transgenic and wild-type plants were observed (Hu et al. 2006), indicating a conserved role of SNAC1 orthologs in drought tolerance in grasses. Under greenhouse conditions, the overexpression of HvSNAC1 in barley plants resulted in higher survival rates and reduced water loss from stressed leaves when compared with wild-type plants. This was reflected in higher RWC, F v/F m and stomatal resistance values of stressed transgenic plants when compared with wild-type plants under the same conditions. These results are in general agreement with previous data on overexpression of stress-responsive NAC genes in different plant species. For instance, the overexpression of HvSNAC1’s highly similar homolog, TaNAC, in Arabidopsis plants resulted in improved plant survival after drought conditions, reduced leaf water loss rate and better photosynthetic capabilities when compared with wild-type plants. Similarly, the overexpression of SNAC1 in rice resulted in increased stomatal closure without affecting the photosynthetic rate. Recently, the characterization of OsSRO1c, which is a direct target of SNAC1 and involved in H2O2-induced stomatal closure, revealed an indirect role of SNAC1 in regulating stomatal closure under stress conditions.
In conclusion, a stress-related NAC transcription factor gene, HvSNAC1 which plays a major role against drought stress in barley was identified and characterized. The overexpression of HvSNAC1 in barley plants resulted in lower water loss rate and yield improvement under drought conditions in the field. The significantly enhanced drought tolerance of the HvSNAC1-overexpressing barley lines indicates its potential for crop improvement in marginal lands.
References
Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9:841–857
Barrs HD, Weatherley PE (1962) A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust J Biol Sci 15:413–428
Bartels D, Sunkars R (2005) Drought and salt tolerance in plants. Crit Rev Plant Sci 24:23–58
Bartlett JG, Alves SC, Smedley M, Snape JW, Harwood WA (2008) High-throughput Agrobacterium-mediated barley transformation. Plant Methods 22:1–12
Bhargava S, Sawant K (2013) Drought stress adaptation: metabolic adjustment and regulation of gene expression. Plant Breed 132:21–32
Choi DW, Rodriguez EM, Close TJ (2002) Barley Cbf3 gene identification, expression pattern, and map location. Plant Physiol 129:1781–1787
Christiansen M, Holm P, Gregersen P (2011) Characterization of barley (Hordeum vulgare L.) NAC transcription factors suggest conserved functions compared to both monocots and dicots. BMC Res Notes 4:302
Collinge M, Boller T (2001) Differential induction of two potato genes, Stprx2 and StNAC, in response to infection by Phytophthora infestans and to wounding. Plant Mol Biol l46:521–526
Delessert C, Kazan K, Wilson I, Straeten D, Manners J, Dennis E, Dolferus R (2005) The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J 43:745–757
Fang Y, You J, Xie K, Xie W, Xiong L (2008) Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Genet Genomics 280:547–563
Habash DZ, Kehel Z, Nachit M (2009) Genomic approaches for designing durum wheat ready for climate change with a focus on drought. J Exp Bot 60:2805–2815
Hegedus D, Yu M, Baldwin D, Gruber M, Sharpe A, Parkin I, Whitwill S, Lydiate D (2003) Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol Biol 53:383–397
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27:297–300
Hu H, Dai J, Yao B, Xiao X, Li Q, Zhang XL (2006) Over-expressing a NAM, ATAF and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103:12987–12992
Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L (2008) Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67:169–181
Jensen M, Rung J, Gregersen P, Gjetting T, Fuglsang A, Hansen M, Joehnk N, Lyngkjaer M, Collinge D (2007) The HvNAC6 transcription factor: a positive regulator of penetration resistance in barley and Arabidopsis. Plant Mol Biol 65:137–150
Jeong J, Kim Y, Baek K, Jung H, Ha S, Choi Y, Kim M, Reuzeau C, Kim J (2010) Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol 153:185–197
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Lu P, Chen N, An R, Su Z, Qi B, Ren F, Chen J, Wang X (2007) A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol Biol 63:289–305
Mao X, Zhang H, Qian X, Li A, Zhao G, Jing R (2012) TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J Exp Bot 63:2933–2946
Maxwell K, Johnson G (2000) Chlorophyll fluorescence: a practical guide. J Exp Bot 51:659–668
Mir RR, Zaman-Allah M, Sreenivasulu N, Trethowan R, Varshney RK (2012) Integrated genomics, physiology and breeding approaches for improving drought tolerance in crops. Theor Appl Genet 125:625–645
Morran S, Eini O, Pyvovarenko T, Parent B, Singh R, Ismagul A, Eliby S, Shirley N, Langridge P, Lopato S (2011) Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol J 9:230–249
Mysore K, Crasta R, Tuori P, Folkerts O, Swirsky B, Martin B (2002) Comprehensive transcript profiling of Pto and Prf-mediated host defense responses to infection by Pseudomonas syringae pv tomato. Plant J 32:299–315
Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51:617–630
Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S (2009) Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signalling are essential for the control of seed development and dormancy. Plant Cell Physiol 50:1345–1363
Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819:97–103
Olsen A, Ernst H, Leggio L, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10:79–87
Puranik S, Sahu P, Srivastava P, Prasad M (2012) NAC proteins: regulation and role in stress tolerance. Trends Plant Sci 17:369–381
Ren T, Qu F, Morris TJ (2000) HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. Plant Cell 12:1917–1926
Saad AS, Li X, Li H, Huang T, Gao C, Guo M, Cheng W, Zhao G, Liao Y (2013) A rice stress-responsive NAC gene enhances tolerance of transgenic wheat to drought and salt stresses. Plant Sci 203–204:33–40
Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K (2001) Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses using full-length cDNA microarray. Plant Cell 13:61–72
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson J, Gibson T, Plewniak F, Jeanmougin F, Higgins D (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tondelli A, Francia E, Barabaschi D, Aprile A, Skinner JS, Stockinger EJ, Stanca M, Pecchioni N (2006) Mapping regulatory genes as candidates for cold and drought stress tolerance in barley. Theor Appl Genet 112:445–454
Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Engineering drought tolerance in plants: discovering and tailoring genes unlock the future. Curr Opin Biotechnol 17:113–122
Vandesompele J, Preter KD, Pattyn F, Poppe B, Roy NV, Paepe AD, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:research0034.1–research0034.11
Wang M, Waterhouse PM (1997) A Rapid and simple method of assaying plants transformed with hygromycin or PPT resistance genes. Plant Mol Biol Rep 15:209–215
Xia N, Zhang G, Liu X, Deng L, Cai G, Zhang Y, Wang X, Zhao J, Huang L, Kang Z (2010) Characterization of a novel wheat NAC transcription factor gene involved in defense response against stripe rust pathogen infection and abiotic stresses. Mol Biol Rep 37:3703–3712
Xue G, Waya H, Richardsonb T, Drentha J, Joycec P, McIntyrea L (2011) Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol Plant 4:697–712
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Bio l57:781–803
Youens-Clark K, Buckler E, Casstevens T, Chen C, DeClerck G, Derwent P, Dharmawardhana P, Jaiswal P, Kersey P, Karthikeyan A, Lu J, McCouch S, Ren L, Spooner W, Stein J, Thomason J, Wei S, Ware D (2010) Gramene database in 2010: updates and extensions. Nucleic Acids Res 39:D1085–D1094
Zhu J (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Zimmermann P, Laule O, Schmitz J, Hruz T, Bleuler S, Gruissem W (2008) GeneVestigator transcriptome meta-analysis and biomarker search using rice and barley gene expression databases. Mol Plant 1:851–857
Acknowledgments
We gratefully acknowledge Mrs. Samar Misbeh, Miss Shireen Qasrawi and Mr. Mohammad Sheik Omar for technical assistance. This work was supported in part by a Grant from the Scientific Research Fund, Ministry of Higher Education, in part by a Grant from the Deanship of Scientific Research, University of Jordan and in part by a Grant from the International Foundation of Science/Sweden.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Al Abdallat, A.M., Ayad, J.Y., Abu Elenein, J.M. et al. Overexpression of the transcription factor HvSNAC1 improves drought tolerance in barley (Hordeum vulgare L.). Mol Breeding 33, 401–414 (2014). https://doi.org/10.1007/s11032-013-9958-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-013-9958-1