Abstract
Two Chinese cabbage (Brassica rapa L. ssp. pekinensis) lines resistant to Turnip mosaic virus (TuMV) CHN5 were identified and found to have broad-spectrum resistance against three other TuMV strains (CHN2, 3, and 4). Genetic analysis indicated that this TuMV resistance is recessive, and a candidate gene approach was used to identify the resistance gene, which we named trs (TuMV resistance discovered at Seoul National University). Based on previous research in Arabidopsis showing that mutations in eIF(iso)4E determine TuMV resistance, the eIF(iso)4E gene was selected as a candidate for the trs gene in Brassica rapa. Three copies of eIF(iso)4E, Braiso4Ea, Braiso4Eb, and Braiso4Ec, were amplified, and polymorphisms between resistant and susceptible lines were analyzed. Sequence polymorphisms were found in Braiso4Ea and Braiso4Eb; in contrast, no sequence differences were found in Braiso4Ec between resistant and susceptible lines. A CAPS marker developed to test the linkage between Braiso4Eb and TuMV resistance displayed no linkage. A SCAR marker, trsSCAR, developed using allele-specific deletions and SNPs in Braiso4Ea, did co-segregate perfectly with trs in three F 2 populations. However, the presence or absence of the Braiso4Ea sequence deletion was not consistent between resistant lines and susceptible lines, indicating that Braiso4Ea is not the actual resistance gene. Results from mapping analysis indicated that the trs is located at chromosome A04, between scaffold 000104 and scaffold 040552. This location demonstrated that trs may be another recessive resistance gene tightly linked to retr02 or another allele. The molecular markers developed in this study will be useful for breeding durable resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Turnip mosaic virus (TuMV), a member of the Potyvirus genus, has a genome consisting of a positive single-stranded RNA molecule of about 10 kb. A viral protein, VPg (22–24 kDa), is covalently linked to the 5′-end of the genome and a poly (A) tail is present at the 3′-end. TuMV infects a wide range of cultivated plants, but the major hosts of the virus are Brassicaceous species, causing severe economic losses in terms of production in Asia, North America, and Europe (Edwardson and Christie 1991; Shattuck et al. 1992). It is aphid transmitted in the non-persistent mode of plant virus transmission (Shattuck et al. 1992). The most common symptom of the virus is a distinct mosaic of light- and dark-green coloration in the leaves. Depending upon the virus strain and the crop species, necrotic streaks, flecks, or ring spots may also appear in the infected crops.
Plant virus resistance genes can be dominant or recessive. Most resistance genes are dominant genes that encode R proteins which usually trigger the hypersensitive response. By contrast, recessive resistance more often occurs due to mutation or loss of host factors. Because plant viruses can encode only a few proteins (about 4–10 proteins), viral infection is highly dependent on the availability of its host factors (Kang et al. 2005), and viral susceptibility is often determined by the host factors. Previous studies have demonstrated that eIF4E protein families are major determinants of recessive resistance against potyviruses (Robaglia and Caranta 2006), with multiple potyvirus recessive resistance genes identified among eIF4E family members, including pvr1 in Capsicum (Ruffel et al. 2002; Kang et al. 2005), mo1 in lettuce (Nicaise et al. 2003), sbm1 in pea (Gao et al. 2004), pot-1 in tomato (Ruffel et al. 2005), and lsp1 resistance alleles created by mutagenesis in Arabidopsis thaliana (Lellis et al. 2002; Yoshii et al. 2004). rym4/5 in barley, which control resistance against Bymoviruses, were also identified as eIF4E family members (Stein et al. 2005), and the retr01 and retr02 TuMV resistance genes are also considered to encode eIF(iso)4E in Chinese cabbage (Rusholme et al. 2007; Qian et al. 2013).
Chinese cabbage, a vegetable, oilseed, and fodder crop, is grown worldwide but especially in Asia and Europe. Despite the serious economic loss of Brassica crops caused by TuMV, there has been no report of TuMV resistance genes that could be deployed stably in resistance breeding. TuMV resistance genes that have been identified in Brassicaceae include TuRB01, TuRB03, TuRB04, and TuRB05 in Brassica napus (Robbins et al. 1994; Walsh et al. 1999; Hughes et al. 2003; Jenner et al. 2002) and TuRB01b, ConTR01, retr01, and retr02 in Brassica rapa (Walsh et al. 2002; Rusholme et al. 2007; Qian et al. 2013). Most of these genes are dominant resistance genes (R genes), with each gene showing a narrow spectrum of resistance to specific TuMV isolates. retr01, however, is a recessive resistance gene that shows broad-spectrum resistance to TuMV strains together with the other dominant gene named ConTR01 (Rusholme et al. 2007). Qian et al. (2013) recently reported for the first time the presence of a single recessive resistance gene, retr02, in Chinese cabbage.
eIF(iso)4E of Arabidopsis thaliana is known to be the essential host factor in TuMV infection (Léonard et al. 2000; Lellis et al. 2002; Duprat et al. 2002; Beauchemin et al. 2007; Miyoshi et al. 2008). Several additional reports have described the relationship between eIF(iso)4E and Brassica recessive resistance. One of the three copies of eIF(iso)4E in Brassica rapa was identified as likely being related to retr01 (Rusholme et al. 2007). In addition, the gene prediction result of retr02 also revealed that the allele is coding eIF(iso)4E protein (Qian et al. 2013).
Here we describe Chinese cabbage lines that display strong resistance to several TuMV strains and the development of a marker for that resistance. Compared to previous reports on recessive resistance of Brassica rapa, the gene that we identified is a single recessive gene that solely controls four different strains of TuMV. Genetic analysis showed that the TuMV resistance is inherited in a recessive manner. Since eIF(iso)4E has been identified as a Potyvirus resistance candidate gene, we analyzed this gene family in more detail.
Materials and methods
Plant materials and virus strains
Brassica rapa subsp. pekinensis ‘Samjin’ (Monsanto Korea, Chochiwon, Korea) was used as a susceptible control and for the cloning of susceptible eIF(iso)4E complementary DNA (cDNA). Chinese cabbage lines (SB15, SB16, SB17, SB18, SB20, SB22, SB23, SB24, and SB25) were provided by the National Institute of Horticultural and Herbal Science and were used for the extraction of eIF(iso)4E genomic DNAs and cDNAs. Another susceptible Chinese cabbage cultivar, ‘GJS2A’, was provided by the Hankook Seed. Co., Ltd. (Ansung, Korea) and used for the construction of F 2 populations. The susceptible Chinese cabbage lines SB20, SB24, and GJS2A and the resistant Chinese cabbage lines SB18 and SB22 were crossed to obtain F 1 plants. F 1 plants were self-pollinated to generate the F 2 populations. For the genetic analysis, we used 57 F 2 individuals from SB22 and SB24 parents, 71 F 2 individuals from SB18 and SB20 parents, and 155 F 2 individuals from GJS2A and SB18 parents.
TuMV CHN2, 3, 4 and 5 were provided by Namhan Huh (Nongwoo Bio, Yeojoo, Korea). Virus inoculum was propagated in susceptible Chinese cabbage cultivar ‘Samjin’. TuMV CHN2, 3, 4 and 5 were used to test the resistance spectrum of Chinese cabbage lines. The TuMV CHN5 strain was used for the F 2 screening.
Virus screening procedure
Plants were inoculated at the two to four leaf stages by mechanical inoculation. Virus inoculum was prepared by grinding TuMV-inoculated leaves in 50 mM potassium phosphate buffer (pH 7.5). Mechanical inoculation was carried out by applying virus inoculum with light carborundum dusting. Plants were monitored daily after 20 days post-inoculation (DPI) and classified as resistant or susceptible to TuMV by the absence or presence of visual virus symptoms, respectively. Susceptibility or resistance was confirmed by the double antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA) using TuMV antibodies (Kisan Biotech Co. Ltd, Seoul, Korea). The plants were scored as susceptible when the ELISA absorbance value was higher than 2.5-fold the mean absorbance value of three uninoculated samples.
Cloning and sequence analysis of eIF(iso)4E
Total RNA was isolated from Chinese cabbage using TRIzol (Invitrogen Life Technologies, Carlsbad, CA). Total genomic DNA was isolated from young green leaves of Chinese cabbage using the CTAB method of Hwang et al. (2009). The full length of eIF(iso)4E sequences were amplified by reverse transcription (RT)-PCR with gene-specific primers [Electronic Supplementary Material (ESM) Table 1]. The PCR assay to amplify cDNAs and genomic DNAs was performed in a total reaction volume of 50 μl containing 50–100 ng of DNA as template, 1× PCR buffer (Takara Shuzo Co., Kyoto, Japan), 2.5 mM dNTP, 1.25 units of EX-Taq (Takara Shuzo Co.), and 5 pmol of each primer. The PCR conditions were 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min. DNA fragments were cut from the gel, and DNA was recovered using a Zymo Gel Recovery kit (Zymo Research, Orange, CA). PCR products were cloned using a TOPO TA Cloning kit (with pCR2.1-TOPO; Invitrogen Life Technologies), and sequencing was performed at NICEM (Seoul National University, Seoul, Korea) by the Sanger method.
For the 3′ amplification of cDNA ends, cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen Life Technologies) using the oligo(dT)-containing a 3′ RACE-AP (rapid amplification of cDNA ends-adapter primer primer; Invitrogen Life Technologies) and an eIF(iso)4E gene-specific primer (ESM Table 1). 3′-RACE fragments were purified and cloned into T-blunt vector (SolGent Co., Daejeon, Korea) and then sequenced.
Phylogenetic analysis of eIF4E family in Brassica was performed using the MEGA software program V5 (Tamura et al. 2007) with 1,000 bootstrap replications.
Developing eIF(iso)4E molecular markers
The eIF(iso)4E gene was used as a TuMV resistance marker in Chinese cabbage. A National Center for Biotechnology Information (NCBI) BLAST nucleotide search was performed in advance. Primers were designed based on eIF(iso)4E genomic DNA sequence alignment data using the Brassica rapa accessions HM131209.1 (BraA. eIF(iso)4E a), HM131211.1 (BraA. eIF(iso)4E c), and HM131210.1 (BraA. eIF(iso)4E b).
For the Braiso4Ea association analysis, a sequence-characterized amplified region (SCAR) marker was developed. Three marker primers were designed based on the sequence alignment of Braiso4Ea-1 and a-2. The PCR analysis was performed in a total reaction volume of 25 μl containing 50–100 ng of genomic DNA as template, 1× PCR buffer [1.56 mM MgCl2, 62.5 mM KCl, 12.5 mM Tris–Cl (PK Science, Seoul, Korea)], 2.5 mM dNTP, 0.2 U of home-made Taq DNA polymerase purified as previously reported (Desai et al. 1995), 6 pmol of primer F 1, 2 pmol of primer F 2, and 5 pmol of primer R (ESM Table 1). The PCR cycling conditions were 37 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min 40 ss. To analyze the co-segregation of Braiso4Eb, we first amplified Braiso4Eb with gene-specific primers and then digested the products with XbaI based on the presence of single nucleotide polymorphisms (SNPs) between susceptible and resistant sequences.
Linkage analysis and resistance gene mapping
The scaffold 000104 and its nine neighborhood scaffolds have previously been mapped at chromosome A04 (Wang et al. 2011; Qian et al. 2013). We designed primers at these ten scaffolds, including scaffold 000104, and sequenced their PCR products. Genomic DNA of SB18, GJS2A, SB22, and SB24 were used for the sequencing, and the sequences were aligned to identify single nucleotide polymorphisms (SNPs) for high-resolution melting (HRM) marker development. Based on the results, we partially sequenced DNA fragment from the ten scaffolds anchored in chromosome A04 to develop SNP markers. SNPs on these scaffolds were identified and marker primers were designed to amplify DNA fragments, including SNPs (ESM Table 1). The HRM method was performed in a reaction volume of 20 μl containing 125 ng of genomic DNA as template, 1× PCR buffer [1.25 mM MgCl2, 50 mM KCl, 10 mM Tris–Cl (PK Science)], 2.5 mM dNTPs 1.25 μM SYTO9 (Invitrogen Life Technologies), 10 pmol of reverse and forward primers, and 0.2 U of home-made Taq DNA polymerase, in a Rotor-gene TM 6000 thermocycler (Corbette Research, Sydney, Australia). The cycling conditions were one cycle of 95 °C for 4 min, followed by 55 cycles of 95 °C for 15 s, 50 °C for 15 s, and 70 °C for 30, with holding at 95 °C for 1 min and at 40 °C for 1 min. HRM was analyzed for each 0.1 °C increment between 70 and 95 °C.
Linkage analysis of the markers developed for the trs mapping study was performed using two F 2 populations, GJS2A × SB18 and SB22 × 24. Linkage between SNP markers and the trs locus was established using Carthagene software (de Givry et al. 2005). The Kosambi function was applied to convert recombination fractions into map distances.
Colony analysis for sorting eIF(iso)4E copies
High-resolution melting analysis was used to screen the colonies containing 3′-RACE product clones. The PCR analysis was performed in a total reaction volume of 20 μl containing template, 1× PCR buffer (1.25 mM MgCl2, 50 mM KCl, 100 mM Tris–Cl), 2.5 mM dNTPs, and 1.25 μM SYTO9 (Invitrogen Life Technologies). Intact bacterial cells were re-suspended in the PCR mixture. Cycling and HRM conditions were one cycle of 95 °C for 4 min, followed by 50 cycles of 95 °C for 15 s, 55 °C for 15 s, and 70 °C for 30 s, with holding at 95 °C for 1 min and at 40 °C for 1 min. HRM was analyzed for each 0.1 °C increment between 70 and 95 °C.
Results
TuMV screening to select virus-resistant Chinese cabbage lines
Chinese cabbage lines (SB15, SB17, SB18, SB20, SB22, SB23, SB24 and SB25) that were presumed to have recessive resistance based on previous reports of the National Institute of Horticultural and Herbal Science were screened for TuMV resistance. Chinese cabbage cultivar ‘Samjin’ was used as a positive control because this cultivar is known to be highly susceptible to TuMV. Virus resistance or susceptibility was assessed by both visual symptoms and ELISA absorbance values. A line was considered susceptible when the inoculated plants showed typical symptoms, such as a mosaic of light- and dark-green coloration, dwarf symptoms, necrosis, and puckering. After 20–30 DPI with TuMV CHN5, obvious typical TuMV symptoms were observed on the upper leaves of cultivar ‘Samjin’ (data not shown). Six Chinese cabbage lines (SB15, SB17, SB20, SB23, SB24, SB25) were classified as susceptible based on their symptoms. Lines SB15, SB17, SB20, and SB24 showed strong mosaic symptoms and other significant TuMV symptoms. By contrast, the symptoms in SB23 and SB25, although obvious, were relatively weak compared to those of the other four susceptible lines. Two lines, SB18 and SB22, showed no visible symptoms on both inoculated and upper leaves. The ELISA values of Chinese cabbage lines SB15, SB17, SB20, and SB24 were similar to those of the ‘Samjin’ positive control. In accordance with their weaker symptoms, the ELISA values of SB23 and SB25 were relatively low compared to those of the positive control and the other four susceptible lines. The ELISA values of two lines, SB18 and SB22, were the same as those of the mock inoculated control (Fig. 1a).
Virus resistance screening of Chinese cabbage lines. a Enzyme-linked immunosorbent assay. SB15, 17, 18, 20, 22, 23, 24, and 25 lines were screened. DAS-ELISA was performed at 25 dpi to test the virus accumulation. b Resistance spectrum of SB18 and SB22. Four TuMV strains, CHN2, 3, 4, and 5 referred as C2, C3, C4, and C5 were used for screening. Mock negative control, Pos positive control line ‘Samjin’. c Pictures of Chinese cabbage plants inoculated with TuMV CHN5. The plants were photographed at 57 dpi. SB24, TuMV-susceptible paternal parent SB24; SB22, TuMV-resistant maternal parent SB22; F 1, susceptible F 1 individual; SB20, TuMV-susceptible paternal parent SB20; SB18, TuMV-resistant maternal parent SB18. GJS2A, TuMV-susceptible paternal parent GJS2A. d Virus accumulation of two resistant parents SB18 and SB22, three susceptible parents SB20, SB24, and GJS2A, and their five F 1 combinations SB18 × SB20, GJS2A × SB18, SB22 × SB24, GJS2A × SB22, and SB18 × SB22. DAS-ELISA was performed at 32 dpi to test the virus accumulation. Error bars represent SD
The resistance spectra of these selected plants were evaluated using TuMV-CHN2, 3, 4, and 5. In comparison to the CHN4 and CHN5 strains, CHN2 and CHN3 induced mild symptoms in susceptible lines and Samjin (data not shown). However, there were no differences between the resistant lines SB18 and SB22, and no symptoms appeared in these plants, even in inoculated leaves. ELISA data also confirmed that these two Chinese cabbage lines were highly resistant to the four TuMV strains (Fig. 1b), with the ELISA values of these two lines being uniformly low following inoculation with any of the four TuMV strains, similar to the mock inoculated control.
Inheritance of resistance to TuMV in F 2 populations
To determine the inheritance pattern of TuMV resistance, we crossed resistant Chinese cabbage lines SB18 and SB22 with susceptible lines SB20, SB24, and GJS2A (TuMV susceptible line provided by Hankook seed. Co. Ltd) to obtain F 1 seeds. F 1 plants were then self-pollinated to generate three F 2 populations, SB18 × 20, GJS2A × SB18, and SB22 × 24. Parental lines and the F 1 and F 2 plants were screened with TuMV strain CHN5. All F 1 plants showed susceptibility, indicating that the resistance gene may be the recessive gene (Fig. 1c, d), but the symptoms were milder than those of the susceptible parents (Fig. 1). Stunting and puckering symptoms were much weaker in F 1 plants than in the susceptible parents (Fig. 1c). A total of 71 F 2 plants of SB18 × 20, 155 F 2 plants of GJS2A × SB18, and 57 plants of SB22 × 24 were screened for the phenotypic segregation analysis. In the SB18 × 20 F 2 population, 63 plants were susceptible and eight plants were resistant (Table 1). The segregation ratio of resistance in the F 2 population deviated greatly from a monogenic ratio of resistant to susceptible plants (χ2 = 7.141; P < 0.05). Subsequent screening of the GJS2A × SB18 F 2 population derived from SB18 revealed that 114 plants were susceptible and 41 plants were resistant. Therefore, the segregation ratio of resistance in this F 2 population fitted with a monogenic 1:3 ratio of resistant to susceptible plants (χ2 = 0.174; P = 0.676) (Table 1). In the SB22 × 24 F 2 population, 43 plants were susceptible and 14 plants were resistant, demonstrating that segregation ratio of resistance in the F 2 population also fitted with a monogenic 1:3 ratio of resistant to susceptible plants (χ2 = 0.006; P = 0.938) (Table 1). Based on these results, we concluded that the resistance of SB18 and SB22 is controlled by a single recessive gene, which we denoted trs (TuMV resistance discovered at Seoul National University).
Allelism test of SB18 and SB22 TuMV resistance
Chinese cabbage lines SB18 and SB22 showed strong resistance against TuMV, with the inheritance data indicating that their resistance genes are a single recessive resistance gene. To confirm the allelism of the resistance genes of SB18 and SB22, we screened resistant × resistant and resistant × susceptible crosses by inoculating five to seven F 1 progenies (SB18 × 22, SB18 × 20, GJS2A × SB18, SB22 × 24, GJS2A × SB22) and their parents with TuMV CHN5. Virus resistance or susceptibility was assessed by both visual symptoms and ELISA absorbance values at 15, 22 and 32 DPI. F 1 SB18 × 20, GJS2A × SB18, SB22 × 24, and GJS2A × SB22 plants showed TuMV susceptibility as expected. By contrast, the F 1 seedlings from the cross between the SB18 and SB22 resistance lines showed uniform resistance (Fig. 1c). Symptoms developed in the parents 7–10 days faster than in the F 1 plants. At 40 DPI, none of the inoculated F 1 plants derived from SB18 × 22 showed visible symptoms, whereas other F 1 plants showed typical TuMV symptoms (ESM Figure 1). The ELISA values confirmed the results (Fig. 1c). These results demonstrate that the TuMV resistance genes of SB18 and SB22 are controlled by the same gene.
DNA sequence analysis of eIF(iso)4E copies
Because mutations in eIF(iso)4E resulted in TuMV resistance in Arabidopsis (Lellis et al. 2002; Yoshii et al. 2004), we chose eIF(iso)4E as a candidate gene for trs. To analyze its genomic DNA sequence, we extracted genomic DNA from resistant and susceptible lines and amplified eIF(iso)4E using several gene specific primers (ESM Table 1).
Cloned genomic DNA was sequenced and three copies of eIF(iso)4E were found. The identified eIF(iso)4E copies were named Braiso4Ea, Braiso4Eb, and Braiso4Ec to distinguish them from the NCBI reference sequences. The copy named Braiso4Eb shows 99 % identity to BraA. eIF(iso)4E b, and the copy named Braiso4Ec has 96 % identity to BraA. eIF(iso)4E c (NCBI BLAST: http://www.ncbi.nlm.nih.gov/BLAST/). There were two alleles of the Braiso4Ea copy, denoted Braiso4Ea-1 and Braiso4Ea-2, and these share 96 and 97 % identity with BraA. eIF(iso)4E a, respectively. However, these two alleles were found to differ greatly in SNPs and 3′ end sequences. We found that SB20 and SB22 have the Braiso4E a-1 allele, whereas SB18 and SB24 have the Braiso4Ea-2 allele (Fig. 2). The other susceptible Chinese cabbage lines (SB15, SB17, SB23, SB25) all have the Braiso4E a-1 allele, similar to SB20 (data not shown). The genome structures of the two Braiso4Ea alleles were found to be quite different, with exons 4 and 5 of Braiso4Ea-1 missing in Braiso4Ea-2. Sequence polymorphisms were observed mostly in introns rather than exons, with the most polymorphic region being the 445- to 545-bp fragment in intron 2. Only a few SNPs were discovered in exons, but none of these SNPs showed consistency in resistant and susceptible lines. For the cDNA and amino acid alignments, Braiso4Ea-2 was found to have an early stop codon in exon 3 compared to Braiso4Ea-1 (ESM Figure 2). Exon 3 of Braiso4Ea-2 is approximately 200 bp longer than that of Braiso4Ea-1 because part of the intron is retained in the former, possibly due to incomplete splicing. As the conserved exon parts (exon 4 and 5) are missing in Braiso4Ea-2, the allele may not be functional. These two alleles of Braiso4Ea were used to perform an association study of TuMV resistance.
Comparison of the Braiso4Ea alleles. a Structure of the genes indicating positions of exons, introns, and generic primers used for marker. Exons are indicated by black boxes and introns by thin lines. The size of each sequence is labeled above the structure. b Sequence alignment of the genomic DNA sequences of Braiso4Ea-1 and Braiso4Ea-2 from SB20, SB22, SB18, and SB24. The trsSCAR marker sites based on allele-specific deletions and SNPs are indicated using gray shadow box. F 1 is based on Braiso4Ea-2 polymorphisms and F 2 is based on Braiso4Ea-1 polymorphisms. R is the common reverse primer for F 1 and F 2. c Linkage analysis of the trsSCAR marker. F 2 SB18 × 20, genotype analysis of SB18 × 20 F 2 using the trsSCAR marker. P1 and P2 refer to SB18 and SB20. F 2 SB22 × 24, genotype analysis of SB22 × 24 F 2 using the trsSCAR marker. P1 and P2 refer to SB22 and SB24, respectively. F 2 GJS2A × SB18, genotype analysis of GJS2A × SB18 F 2 using the trsSCAR marker. P1 and P2 refer to SB18 and GJS2A, respectively. M DNA marker, R resistant phenotype, S susceptible phenotype
Following alignment of the Braiso4Eb from each line (ESM Figure 3), we detected SNPs between susceptible and resistant Braiso4Eb copies. These SNPs were used for further analysis of TuMV resistance association. By contrast, there were no sequence variations in Braiso4Ec sequences among two susceptible and two resistant Chinese cabbage lines.
TuMV resistance linkage analysis of Braiso4Ea and Braiso4Eb
Linkage analysis of each eIF(iso)4E copy was performed to test which eIF(iso)4E copy is related to the recessive resistance. As Braiso4Ec did not show specific sequence variation among the four susceptible and two resistant Chinese cabbage lines, only Braiso4Ea and Braiso4Eb copies were tested (ESM Figure 3B).
A SCAR marker was developed for the Braiso4Ea linkage analysis. The primer set was prepared by sequence comparison of the Braiso4Ea-1 and a-2 alleles (Fig. 2), with primer F 1 designed using the specific insertion sequence of SB18 and SB24 located in intron 1 and primer F 2 based on the specific insertion sequence of SB20 and SB22 located in exon 3. The reverse primer R was derived from the sequence located in intron 3 of Braiso4Ea-1, which is shared with the 3′ end of Braiso4E a-2. The F 1 and R primer set can amplify only Braiso4Ea-2 in SB18 and SB24, and the expected PCR product size is about 680 bp, while the F 2 and R primer set can amplify only Braiso4Ea-1 in SB20, SB22, and GJS2A, and the expected PCR product size is about 190 bp. This marker was named the trsSCAR marker (Fig. 2c).
To examine whether the marker could be used for eIF(iso)4E genotyping of the F 2 population, parental lines, the F 1 plants, and F 2 populations of SB18 × 20, GJS2A × SB18, and SB22 × 24 were screened with the trsSCAR marker. Comparison of the band pattern in two resistant (SB18 and SB22) and three susceptible parent lines (SB20, SB24 and GJS2A) and in their F 1 plants (SB18 × 20, GJS2A × SB18, SB22 × 24) revealed polymorphism as expected (Fig. 2c). Among the genomic DNAs from 71 F 2 population individuals from the SB18 × 20 cross, 18 F 2 plants showed a band pattern corresponding to that of the susceptible parental line SB20, 45 plants were confirmed as heterozygous, and the other eight plants showed the same band pattern as the resistant parental line SB18 (Table 2). Screening of the genomic DNAs from 155 GJS2A × SB18 F 2 individuals with the trsSCAR marker that these plants had the same band pattern as SB18 × 20 F 2 plants. Among these genomic DNAs, 38 F 2 plants showed the band pattern corresponding to the susceptible parental line GJS2A, 76 plants were heterozygous, and 41 plants showed the same band pattern as the resistant parental line SB18 (Table 2). Among genomic DNAs from 57 F 2 population individuals from the SB22 × 24 cross, 18 F 2 plants showed the same band pattern as the susceptible parental line SB24, 25 plants were heterozygous, and the other 14 plants showed the same band pattern as the resistant parental line SB22 (Table 2). Plants having the resistant parent genotype showed resistance to TuMV, whereas the plants with the susceptible parent genotype and heterozygous exhibited susceptibility. The distribution of the three genotypes (TRS/TRS, TRS/trs, trs/trs) in the F 2 progeny of SB22 × 24 nearly fitted the 1:2:1 ratio (χ2 = 1.421, P = 0.491). However, the ratio of the three genotypes in the SB18 × 20 F 2 progeny was 2.4:5.8:1 and did not fit with the expected 1:2:1 ratio (χ2 = 7.901, P < 0.05) (Table 2), possibly due to the small F 2 population, which can lead to biased results. Analysis of the GJS2A × SB18 F 2 population showed that distribution of the three genotypes (TRS/TRS, TRS/trs, trs/trs) nearly fitted the 1:2:1 ratio (χ2 = 0.174, P = 0.917).
Linkage analysis of Braiso4Eb was performed using primers designed with the 5′ and 3′ untranslated region (UTR) sequences in order to specifically detect Braiso4Eb. A cleaved amplified polymorphic sequence (CAPS) marker using an XbaI recognition site was developed based on sequence comparison between the resistant and susceptible lines (ESM Figure 3). The susceptible copy has an XbaI restriction enzyme site and is digested into two bands of 1,020 and 262 bp, respectively (ESM Figure 3). The resistant copy does not have an XbaI site. The F 2 population was screened using the marker, and the results showed that the F 2 resistance phenotype did not match the band pattern of the susceptible and resistant parents. The same results were obtained for the SB22 × 24 F 2 population (data not shown). These results indicate that the TuMV resistance in SB18 and SB22 is controlled by the same single recessive gene.
Mapping the trs locus
As the sequence analysis of Braiso4Ea showed that Braiso4Ea is not the gene that controls the TuMV resistance, we searched for other possible candidate genes. BLAST searches using the Brassica database (BRAD: http://brassicadb.org/brad) (ESM Table 2) were initially performed to obtain sequences around Braiso4Ea. This search revealed that all of the homologues of eIF4E family in Brassica rapa had been isolated previously in the Brassica database. The phylogenetic analysis was conducted using MEGA program ver. 5 (Tamura et al. 2007), and those homologues were classified into three distinct groups, namely, eIF4E, eIF(iso)4E, and nCBP (ESM Figure 4). This analysis is based on genomic DNA data, so there may be pseudogenes that are not expressed. The data analysis showed that multiple copies of eIF4E and eIF(iso)4E exist in Brassica rapa. Among several eIF(iso)4E homologues, we found that Braiso4Ea seems to be contained in scaffold 000104 (Bra035393).
Preliminary mapping results using the VCS40 doubled haploid (DH) population showed that trs gene might be located at chromosome A01 or A09 (data not shown). However, further efforts to map the precise position of the trs gene using A01 or A09 markers showed that none of the developed markers were linked to the trs gene, indicating that our initial mapping of the trs position was wrong. Qian et al. (2013) recently reported the possibility that another single recessive TuMV resistance gene named retr02 encodes eIF(iso)4E (Qian et al. 2013). According to their study, retr02 is located on Brassica scaffold 000104 on chromosome A04 (Qian et al. 2013). We suspected that our gene trs may also be located on chromosome A04, not on chromosome A09 where we previously mapped trs.
There are ten scaffolds mapped in the chromosome A04, including scaffold 000104 (Qian et al. 2013). To determine which scaffold is linked to the trs resistance gene, we developed markers from these scaffolds. Those markers showing clear polymorphism were mapped in two F 2 populations (SB22 × 24 and GJS2A × SB18). Markers from four scaffolds (scaffolds 000104, 000083, 040552, 000177) showed linkage between the trs gene (Fig. 3). In the SB22 × 24 F 2 population, four markers showed linkage between the trs gene, namely, Sca104 10-5 HRM (754.7 kb, scaffold 000104), Sca104-1 HRM (543.2 kb, scaffold 000104), Sca83 1-2 HRM (56.2 kb, scaffold 000083), and Sca552 2-8 HRM (403.2 kb, scaffold 040552) (ESM Table 1). All of these four markers co-segregated perfectly with trs resistance in the SB22 × 24 F 2 population. Five markers were mapped in the GJS2A × SB18 F 2 population (ESM Table 1; Fig. 3). Among those markers only the Sca104 4-2 HRM marker and Sca83 1-7 HRM marker co-segregated perfectly with trs resistance (Fig. 3). The Sca104-5 HRM marker developed at the lower end of scaffold 000104 had three recombinants in the Sca552 1-2 HRM marker and one recombinant in the GJS2A × SB18 population (Fig. 3). Based on these results, we assumed that the locus of the trs is between Sca104-5 HRM (12.4 kb, scaffold 000104) and Sca552 1-2 HRM (28.2 kb, scaffold 040552) on chromosome A04 (Fig. 3).
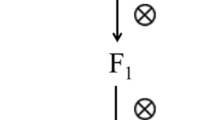
Genetic linkage map of trs locus. GJS2A × SB18 F 2 is the mapping population. Genetic distance in cM was calculated using the Kosambi function. The physical map located on the left side shows the ten scaffolds mapped in chromosome A04 (Qian et al. 2013). The resistance gene trs is located between scaffold 000104 and scaffold 000083
Identification of expressed genes of the eIF4E family
To test whether the candidate eIF4E/eIF(iso)4E genes are expressed, we searched an expressed sequence tag (EST) sequence database (BrTED: http://brted.rna.kr; Yu et al. 2011) (ESM Table 2) for expression of eIF4E family members. We found that eIF4E, eIF(iso)4E, and nCBP were expressed in Brassica rapa and that there were 15–20 EST sequences for each member (data not shown). Among the eIF(iso)4E members, EST sequences for Braiso4Ea and Braiso4Ec were identified. However, there was no sequence that showed similarity to Braiso4Eb, indicating that this copy is not expressed in Brassica rapa. To confirm this result, 3′-RACE PCR was performed using total RNAs of SB18, SB20, SB22, and SB24. The cDNA sequences of two eIF(iso)4E copies, Braiso4Ea and Braiso4Ec, were amplified and the expression levels between two copies were similar (ESM Figure 6). No products corresponding to Braiso4Eb were amplified via 3′-RACE PCR, which is in agreement with the EST data. Also, no other sequences belonging to the eIF4E family other than Bra035393 have to date been found to be expressed in chromosome A04 (ESM Table 2).
Discussion
In this study, we identified broad-spectrum TuMV resistance inherited in a recessive manner. The trsSCAR marker was developed based on two Braiso4Ea alleles. Whereas the trsSCAR marker was observed to co-segregate with the TuMV resistance, two resistant lines showed opposite genotypes for the marker. This result means that Braiso4Ea is tightly linked to the TuMV resistance, but that it is not the trs gene. The allelism test showed that the resistance gene in the SB18 and SB22 lines is the same trs gene, while the mapping result indicates that the trs locus is between scaffold 000104 and scaffold 040552 of chromosome A04.
The first recessive resistance gene found in Chinese cabbage is retr01 (Rusholme et al. 2007). This gene is epistatic to the second dominant gene, Contro1, and Rusholme et al. (2007) reported that these genes may be located in the upper portions of chromosome 4 and 8. These authors also suggested that Contro1 and retr01 may be related to the eIF4E family, but there has been no other report showing the actual relationship between eIF4E family gene and virus resistance in Brassica rapa.
Qian et al. (2013) recently identified a single recessive resistance gene, retr02, in Brassica rapa. These authors speculated that Bra035393 [BraA. eIF(iso)4E a], which is located on scaffold 000104, is the retr02 gene. Our mapping result suggests that the trs locus is located between scaffold 000104 (12.4 kb) and scaffold 040552 (28.2 kb) of chromosome A04 (Fig. 3). Alignment of the DNA sequence of Bra035393 and Braiso4Ea-1 (data not shown) revealed that the Braiso4Ea-1 sequence had 100 % similarity with that of Bra035393. According to our research, however, the presence or absence of deletions in the Braiso4Ea sequence was not consistent between resistant and susceptible Chinese cabbage lines. Therefore even though Braiso4Ea is tightly linked to the trs gene, it is not actually the trs gene itself. Based on this evidence, we conclude that the retr02 and trs genes should be distinguishable, but must point out that the trs locus we mapped overlaps with the locus of retr02. The retr02 locus was mapped on both scaffold 000060 and scaffold 000104. The candidate gene approach showed that retr02 is on scaffold 000104. The trs was mapped between scaffold 000104 and scaffold 040552. Because the identification of the trs gene is still ongoing, we should not rule out that trs may be another recessive resistance gene tightly linked to allele of retr02 or another allele.
To date no other eIF4E family member has been found on chromosome A04. However, the full sequence of chromosome A04 is as yet unknown because of the gaps between scaffolds due to the constraints of next-generation sequencing (NGS) and the presence of abundant repeats in the Brassica rapa genome. Therefore, we cannot rule out the possibility that the trs gene may be located in the gap between scaffold000104 and scaffold040552.
As Braiso4Ea was found to be linked only to trs, we needed to identify other candidates for the trs gene. Other eIF4E or eIF(iso)4E copies may exist in addition to the sequences found in this study. Several similar copies of eIF4E and eIF(iso)4E were found in the Brassica database, and these copies are expected to show redundancy to each other. The presence of these copies indicates that eIF4E family proteins may play different roles in different tissues and/or at different developmental stages in a single organism (Duprat et al. 2002; Rhoads et al. 2007). We found a few cases in which eIF4E or eIF(iso)4E sequences are located very near each other in the same chromosome. On chromosome A8, we found that two eIF(iso)4E sequences (Bra 035531 and Bra 031530) are present within a physical distance of about 2,930 bp. In chromosome A9, two eIF4E sequences (Bra 032325 and Bra 032326) were found to be close, but these were subsequently identified as pseudogenes. Furthermore, we cannot dismiss the possibility that the trs gene may not belong to the eIF4E family.
It was also intriguing that EST sequences showing similarity to the Braiso4Ea copy tend to have many polymorphisms in their open reading frame (ORF) sequences compared to the other copies in the eIF4E family. For example, the sequence named EX042260 (256 bp) had six SNPs in its ORF and CV546692 (440 bp) had 11 SNPs. By contrast, Braiso4Ec copies and other eIF4E groups showed one to three SNPs on average. According to these data eIF4E copy sequences would appear to be highly conserved compared to eIF(iso)4E copies.
It was challenging to analyze the Brassica sequences because there are several known eIF(iso)4E copies. This difficulty in sequence analysis may arise due to the complexity of the Brassica genome. The Brassica rapa genome is organized into ten chromosomes and has emerged as an important model for genomic studies in Brassica species. Comparative studies of Arabidopsis and Brassica prove the occurrence of extensive duplications, and Arabidopsis segments have been conserved an average of three times within the diploid Brassica genomes (Lukens et al. 2003; Parkin et al. 2005). The widespread repetitive sequences in the Brassica genome add to the difficulty in analysis. In the case of eIF(iso)4E, there is only a single copy of the eIF(iso)4E gene in Arabidopsis whereas Brassica rapa possesses multiple copies, including BraA. eIF(iso)4E a. and BraA. eIF(iso)4E c. (confirmed by Jenner et al. 2010).
The trs gene that we found in our study confers a high level of resistance to several TuMV strains, namely, CHN2, 3, 4, and 5. Recessive resistance is known to be more durable and provides broad-spectrum resistance (Kang et al. 2005, 2007). In barley, resistance to the powdery mildew fungus provided by the mlo locus is a typical case of broad-spectrum recessive resistance (Büschges et al. 1997). The pvr1 locus from C. chinense is also famous for its broad-spectrum recessive resistance to several potyviruses, such as Potato virus Y (PVY) pathotypes 0, 1, and 2, Pepper mottle virus (PepMoV), and most Tobacco etch virus (TEV) strains (Kyle and Palloix 1997). pot-1, a recessive resistance gene in tomato, mediates resistance to both TEV and PVY (Parrella et al. 2002; Moury et al. 2004). rym4/5 is also known for controlling the resistance of Barley yellow mosaic virus and Barley mild mosaic virus (Stein et al. 2005). Several sources of broad-spectrum resistance have also been described in Brassica rapa (Yoon et al. 1993; Suh et al. 1995; Liu et al. 1996; Hughes et al. 2002; Walsh et al. 2002; Rosholme et al. 2007). It would appear that broad-spectrum resistance in Brassica tends to be controlled by recessive genes, but the genetic characterization of the recessive resistance was unclear in many cases. Brassica rapa resistance line 0–2 is effective against five TuMV strains (CHN1–5) and has been reported to be controlled by two recessive genes when tested by strains CHN4 and 5 (Yoon et al. 1993). Another resistance line, RLR22, contains two resistance genes, the recessive gene retr01 and the dominant gene Contro1. It also was found to exhibit broad-spectrum resistance against eight different TuMV strains (Rusholme et al. 2007).
Based on our results, the trs gene is inherited as a single recessive gene. However, unlike that of the SB22 × 24 population, the genetic ratio of the SB18 × 20 population did not fit to an expected Mendelian ratio and was approximately 1:8 (resistant vs. susceptible) according to our data. This result can be ascribed to the small size of the F 2 population. Another SB18-derived F 2 population (GJS2A × SB18) did show an expected ratio of 1:3 (resistance vs. susceptible).
The symptoms of the F 1 Chinese cabbage lines were different from those of the susceptible parent even though the virus coat protein accumulations were similar. The virus symptoms of the F 1 plants were much weaker than those of the susceptible parents. The same trend was observed in homozygous resistant compared to heterozygous susceptible F 2 individuals. The symptoms caused by viral systemic infection vary due to differences in host factor–virus interactions (Kaneko et al. 2004; Kim et al. 2010). Therefore, even though host proteins are controlled by recessive alleles, it would seem that the differences in host proteins produced in homozygous and heterozygous plants affect the development of virus symptoms. Previous studies on overexpression of recessive resistance alleles in tomato and potato systems provide evidence for this notion. Kang et al. (2007) reported that the ectopic expression of mutated eIF4E of pepper induced strong resistance to multiple viral species in the tomato system. Another recent study showed that transgenic expression of the pvr1 2 gene from pepper confers resistance to PVY in potato (Cavatorta et al. 2011). Overexpressed eIF(iso)4E mutant protein overwhelmed the normal eIF(iso)4E and the virus–host interaction was disturbed. These reports are consistent with our observation of weak symptoms in the heterozygous plants.
The broad-spectrum resistance reported in this paper represents a novel and potentially durable source of resistance to TuMV. Because this resistance is extremely strong and is controlled by only a single gene, this TuMV resistance source may be useful in developing resistant cultivars, especially in Korea and China where the damage caused by TuMV is severe. The newly developed SCAR markers can be used in marker-assisted selection for TuMV resistance. Because the SCAR marker we developed is based on the copy linked to the trs gene, the trs gene itself should be identified in order to utilize the resistance more efficiently. Furthermore, identification of the trs gene will elucidate fundamental mechanisms of host–virus interaction in Brassica rapa and contribute to a better understanding of virus resistance in plants.
References
Beauchemin C, Boutet N, Laliberté JF (2007) Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of Turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in Planta. J virol 81(2):775–782
Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88:695–705
Cavatorta J, Perez KW, Gray SM, Van Eck J, Yeam I, Jahn M (2011) Engineering virus resistance using a modified potato gene. Plant Biotechnol J 9:1014–1021
de Givry S, Bouchez M, Chabrier P, Milan D, Schiex T (2005) Cartha gene: multipopulation integrated genetic and radiation hybrid mapping. Bioinformatics 21:1703–1704
Desai UJ, Pfaffle PK (1995) Single-step purification of a thermostable DNA polymerase expressed in Escherichia coli. Biotechniques 19(5):780–782
Duprat A, Caranta C, Revers F, Menand B, Browning KS, Robaglia C (2002) The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J 32:927–934
Edwardson JR, Christie RG (1991) Potyviruses. Fla Agric Exp Stat Monogr Ser 16, vol 1–4
Gao Z, Eyers S, Thomas C, Ellis N, Maule A (2004) Identification of markers tightly linked to sbm recessive genes for resistance to Pea seed-borne mosaic virus. Theor Appl Genet 109(3):488–494
Hughes S, Green S, Lydiate D, Walsh J (2002) Resistance to Turnip mosaic virus in Brassica rapa and B. napus and the analysis of genetic inheritance in selected lines. Plant Pathol 51:567–573
Hwang J, Li J, Liu WY, An SJ, Cho H, Her NH, Yeam I, Kim D, Kang BC (2009) Double mutations in eIF4E and eIFiso4E confer recessive resistance to Chilli veinal mottle virus in pepper. Mol Cells 27:329–336
Jenner CE, Nellist CF, Barker GC, Walsh JA (2010) Turnip mosaic virus (TuMV) is able to use alleles of both eIF4E and eIF(iso)4E from multiple loci of the diploid Brassica rapa. Mol Plant Microbe Interact 23:1498–1505
Kaneko Y, Inukai T, Suehiro N, Natsuaki T, Masuta C (2004) Fine genetic mapping of the TuNI locus causing systemic veinal necrosis by Turnip mosaic virus infection in Arabidopsis thaliana. Theor Appl Genet 110:33–40
Kang BC, Yeam I, Jahn MM (2005) Genetics of plant virus resistance. Annu Rev Phytopathol 43:581–621
Kang BC, Yeam I, Li H, Perez KW, Jahn MM (2007) Ectopic expression of a recessive resistance gene generates dominant potyvirus resistance in plants. Plant Biotechnol J 5:526–536
Kim BM, Suehiro N, Natsuaki T, Inukai T, Masuta C (2010) The P3 protein of Turnip mosaic virus can alone induce hypersensitive response-like cell death in Arabidopsis thaliana carrying TuNI. Mol Plant Microbe Interact 23:144–152
Kyle M, Palloix A (1997) Proposed revision of nomenclature for potyvirus resistance genes in Capsicum. Euphytica 97:183–188
Lellis AD, Kasschau KD, Whitham SA, Carrington JC (2002) Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr Biol 12:1046–1051
Léonard S, Plante D, Wittmann S, Daigneault N, Fortin MG, Laliberté JF (2000) Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J Virol 74(17):7730–7737
Liu X, Lu W, Liu Y, Wei S, Xu J, Liu Z, Zhang H, Li J, Ke G, Yao W (1996) Occurrence and strain differentiation of turnip mosaic potyvirus and sources of resistance in Chinese cabbage in China. Acta Hort 407:431–440
Lukens L, Zou F, Lydiate D, Parkin I, Osborn T (2003) Comparison of a Brassica oleracea genetic map with the genome of Arabidopsis thaliana. Genetics 164:359–372
Miyoshi H, Okade H, Muto S, Suehiro N, Nakashima H, Tomoo K, Natsuaki T (2008) Turnip mosaic virus VPg interacts with Arabidopsis thaliana eIF(iso)4E and inhibits in vitro translation. Biochimie 90(10):1427–1434
Moury B, Morel C, Johansen E, Guilbaud L, Souche S, Ayme V, Caranta C, Palloix A, Jacquemond M (2004) Mutations in Potato virus Y genome-linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum. Mol Plant Microbe Interact 17:322–329
Nicaise V, German-Retana S, Sanjuán R, Dubrana MP, Mazier M, Maisonneuve B, LeGall O (2003) The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus Lettuce mosaic virus. Plant Physiol 132(3):1272–1282
Parkin IAP, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ (2005) Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171:765
Parrella G, Ruffel S, Moretti A, Morel C, Palloix A, Caranta C (2002) Recessive resistance genes against potyviruses are localized in colinear genomic regions of the tomato (Lycopersicon spp.) and pepper (Capsicum spp.) genomes. Theor Appl Genet 105:855–861
Qian W, Zhang S, Zhang S, Li F, Zhang H, Wu J, Wang X, Walsh JA, Sun R (2013) Mapping and candidate-gene screening of the novel Turnip mosaic virus resistance gene retr02 in Chinese cabbage (Brassica rapa L.). Theor Appl Genet 126:179–188
Rhoads RE, Dinkova TD, Jagus R (2007) Approaches for analyzing the differential activities and functions of eIF4E family members. Method Enzymol 429:261–297
Robaglia C, Caranta C (2006) Translation initiation factors: a weak link in plant RNA virus infection. Trends in Plant Science 11(1):40–45
Ruffel S, Dussault MH, Palloix A, Moury B, Bendahmane A, Robaglia C, Caranta C (2002) A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J 32(6):1067–1075
Ruffel S, Gallois JL, Lesage ML, Caranta C (2005) The recessive potyvirus resistance gene pot-1 is the tomato orthologue of the pepper pvr2-eIF4E gene. Mol Genet Genomics 274(4):346–353
Rusholme RL, Higgins EE, Walsh JA, Lydiate DJ (2007) Genetic control of broad-spectrum resistance to turnip mosaic virus in Brassica rapa (Chinese cabbage). J Gen Virol 88:3177–3186
Shattuck VI (1992) The biology, epidemiology, and control of turnip mosaic virus. Hortic Rev 14:199–238
Stein N, Perovic D, Kumlehn J, Pellio B, Stracke S, Streng S, Ordon F, Graner A (2005) The eukaryotic translation initiation factor 4E confers multiallelic recessive Bymovirus resistance in Hordeum vulgare (L.). Plant J 42:912–922
Suh S, Green S, Park H (1995) Genetics of resistance to five strains of turnip mosaic virus in Chinese cabbage. Euphytica 81:71–77
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Walsh JA, Rusholme RL, Hughes SL, Jenner CE, Bambridge JM, Lydiate DJ, Green SK (2002) Different classes of resistance to turnip mosaic virus in Brassica rapa. Eur J Plant Pathol 108:15–20
Wang Y, Sun S, Liu B, Wang H, Deng J, Liao Y, Wang Q, Cheng F, Wang X, Wu J (2011) A sequence-based genetic linkage map as a reference for Brassica rapa pseudochromosome assembly. BMC Genomics 12:239
Yoon J, Green S, Opena R (1993) Inheritance of resistance to turnip mosaic virus in Chinese cabbage. Euphytica 69:103–108
Yoshii M, Nishikiori M, Tomita K, Yoshioka N, Kozuka R, Naito S, Ishikawa M (2004) The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J Virol 78:6102
Yu HJ, Park SG, Oh M, Hwang HJ, Kim N, Chung H, Sohn SH, Park BS, Mun JH (2011) The Brassica rapa tissue-specific EST database. Korean J Hort Sci Technol 29:633–640
Acknowledgments
This study was supported by a Grant (Project No. 609002-5) from the Screening Center for Disease Resistance Vegetable Crops of Technology Development Program, Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea, a Grant (Project No. 710001-03) from the Vegetable Breeding Research Center through the R&D Convergence Center Support Program, Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea, and from the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2010-0005936).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, J., Kang, WH., Yang, HB. et al. Identification of a broad-spectrum recessive gene in Brassica rapa and molecular analysis of the eIF4E gene family to develop molecular markers. Mol Breeding 32, 385–398 (2013). https://doi.org/10.1007/s11032-013-9878-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-013-9878-0