Abstract
Quantitative real-time polymerase chain reaction (qRT-PCR) has been extensively used in several plant species as an accurate technique for gene expression analysis. However, the expression level of a target gene may be misconstrued due to unstable expression of the reference genes under different experimental conditions. Therefore, it is necessary to systematically evaluate these reference genes before experiments are conducted. Recently, more and more studies have focused on gene expression in pepper (Capsicum annuum L.). In this study, ten putative reference genes were chosen to identify expression stability by using geNorm and NormFinder statistical algorithms in ten different pepper sample pools, including those from different plant tissues (root, stem, leaf and flower) and from plants treated with hormones (salicylic acid and gibberellic acid) and abiotic stresses (cold, heat, salt and drought). EF1α and UEP exhibited the most stable expression across all of the tested pepper samples. For abiotic stress or different hormone treatment, the ranking of candidate reference genes was not completely consistent, except for EF1α which showed a relatively stable expression level. For different tissues, the expression of Actin1 was stable and it was considered an appropriate reference gene. It is concluded that EF1α, UEP and Actin1 are suitable reference genes for reliable qRT-PCR data normalization for the tissues and experimental conditions used in this experiment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Due to its high sensitivity and specificity, real-time quantitative (qRT-PCR) has become one of the most widely used methods for gene expression analysis in a number of biological areas, including biotechnology, microbiology, and the diagnosis of infections (Bustin et al. 2005; Ginzinger 2002; Giulietti et al. 2001; Weis et al. 1992; Gachon et al. 2004). However, many variables must be considered when using qRT-PCR to analyze gene expression, including the amount of starting material, RNA quality, cDNA sample loading, amplification efficiencies, and the selection of endogenous reference genes. Of these, the use of suitable reference genes for the normalization of gene expression is an elementary prerequisite for reliable results in any qRT-PCR analysis.
Generally, a suitable reference gene for gene expression analysis should be defined as a gene which is stably expressed among the different samples used and unaffected by any experimental treatment. Meanwhile, it should not be associated with any pseudogene, to avoid genomic DNA amplification, while its stability should be equivalent to that of the target gene transcript (Lland et al. 2006). Currently, many kinds of endogenous reference genes such as tubulin (alpha- or beta-), ribosomal units (18 or 28 s rRNA), and ubiquitin (UBQ) (Huggett et al. 2005; Yusuke et al. 2006) are widely used to normalize data in gene expression analysis. However, a large number of recent studies have demonstrated that some of the most well-known and frequently used reference genes are inappropriate for normalization in qRT-PCR analysis due to expression variability (Czechowski et al. 2005; Jarošova and Kundu 2010; Remans et al. 2008; Schmittgen and Zakrajsek 2000). Theoretically, reference genes should be essential for the maintenance of cellular function and vitality and should be stably expressed in tissues and cells, but in practice their expression levels are dependent on tissue types, developmental stages, organisms, and experimental conditions (Yoo et al. 2009; Sturzenbaum and Kille 2001). Furthermore, no single reference gene can be used under different experimental conditions. It is also known that single reference genes are inadequate for normalization purposes in gene expression analyses. Thus, it is necessary to perform systematic validation of reference genes as an essential component of qRT-PCR analysis to improve the reliability of results.
Recently, several statistical analysis methods, such as geNorm (Vandesompele et al. 2002) and NormFinder (Andersen et al. 2004), have been proposed for evaluating the expression stability of reference genes and selecting the most suitable reference genes. These methods are based on different statistical algorithms, using multiple reference genes as the best strategy for normalization of qRT-PCR results. The geNorm applet provides a measure of gene expression stability (M), which is the mean pair-wise variation between an individual gene and all other tested control genes (Vandesompele et al. 2002). The most and least stable reference genes are indicated by lowest and highest values of M, respectively. In addition, geNorm performs a stepwise exclusion of the gene with the highest M value, resulting in the best combination of two reference genes with the most stable expression profiles. The NormFinder approach focuses on finding the two genes with smallest intra- and inter-group expression variation, where groups are defined as different experimental conditions (Andersen et al. 2004). Since the stability value is a combination of intra- and inter-group variation parameters, NormFinder ranks the genes that present minimal variation instead of candidates with correlated expression.
Hot pepper (Capsicum annuum L.) is one of the most important vegetables worldwide. Recently, more studies have focused on its gene expression (Kim et al. 2007; Dilger et al. 2003; Narasimha et al. 2009). However, there are very few studies of gene expression analysis using qRT-PCR in pepper or on appropriate reference gene selection in this species. The aim of this study was to evaluate appropriate reference genes for qRT-PCR gene expression studies in pepper. We evaluated ten pepper candidate reference genes (Actin, Actin1, Actin2, 18S rRNA, PPR1, GAPDH, TEF1A, EF1α, UEP, and CYC) and their expression stabilities were subsequently tested in two hormone treatments (salicylic acid and gibberellic acid), four abiotic stress treatments (cold, heat, salt, and drought), and four different pepper tissues (root, stem, leaf, and flower) using geNorm and NormFinder.
Materials and methods
Plant material
CMS 21A, a strict cytoplasmic male sterile pepper line, was studied (Zhao et al. 1995). In the spring of 2009, seeds of CMS 21A were germinated and grown in the greenhouse for 12 h light at 30 °C and 12 h dark at 20 °C. The relative humidity was kept at 65–75 %.
Abiotic stress treatments
For salt- and drought-stress treatments, pre-florescence seedlings were transferred into pots containing 300 mM NaCl or 300 mM mannitol for 12 h. For cold- and heat-stress treatments, the seedlings were kept for 12 h at 4 ± 1 °C and 42 ± 1 °C, respectively. The seedlings that were kept in water for the same duration at 25 ± 1 °C served as control. After the prescribed treatment duration, leaves were harvested for RNA extraction.
Hormone treatments
The seedlings at the four true-leaf stage were sprayed using solutions of salicylic acid (SA, 100 μM) and gibberellic acid (GA, 100 μM), respectively, and the leaves sampled after 10 h. Plants were sprayed with sterile water for control.
Different tissues
Roots, stems, leaves, and flowers were harvested from CMS 21A in florescence. All samples were immediately frozen in liquid nitrogen, and stored at −70 °C until needed for RNA extraction.
Total RNA isolation and cDNA synthesis
Total RNA was isolated from the roots, stems, leaves, and flowers of CMS 21A using RNAsimple total RNA kit (Takara, Japan) and was treated with DNase I (Takara) to remove any traces of genomic DNA, according to the manufacturer’s instructions. RNA concentration and purity were determined using a NanoDropTM spectrophotometer ND-1000 (Thermo Scientific), and RNA integrity was verified in 1 % agarose gel electrophoresis.
The first-strand cDNA was synthesized by adding 2 μl of 10 μM random primer, 2 μl of 2.5 mM dNTPs, and 2.5 μg of total RNA to sterile distilled water in a total volume of 13.5 μl. This mixture was incubated at 70 °C for 5 min and briefly chilled in ice for 2 min. Then 4 μl 5 × First Strand Buffer, 1 μl 0.1 M dithiothreitol (DTT), 0.5 μl 20 U/μl RNasin, and 1 μl 200 U/μl TIANScript M-MLV were added to the previous mixture and incubated at 25 °C for 10 min, then at 42 °C for 50 min. The reverse transcriptase was inactivated to stop the reaction at 95 °C for 5 min, following the manufacturer’s instructions. RNA extraction and cDNA synthesis from all of the different samples were performed for two biological replicates.
Primer design, qRT-PCR, and data analysis
A total of ten pairs of specific primers for reference genes were designed using Beacon Designer 2.06 (Primer Biosoft International), and details of these primers are provided in Table 1. qRT-PCR reaction was carried out in a total volume of 25 μl containing 12.5 μl 2 × SYBR Premix Ex Taq™ (Takara), 1 μl 10 pM of each primer, 2 μl template (10 × diluted cDNA from samples), and 8.5 μl sterile distilled water. Reaction mixtures were incubated for 30 s at 95 °C, followed by 40 amplification cycles for 5 s at 95 °C and 30 s at 60 °C. All reactions were carried out on 96-well reaction plates with the iQ5 machine (Bio-Rad) in triplicate. qRT-PCR analysis was performed by the comparative Ct method, which mathematically transforms the threshold cycle (Ct) into the relative expression level of genes (Perkin-Elmer User Bulletin). Primer efficiencies were calculated with LinRegPCR v11.0 software. Relative expression levels of these genes were imported to geNorm v3.5 and NormFinder analysis tools, which were used as described in their manuals. Data of three biological replicates were analyzed separately in the two programs.
Results
High variation in expression levels of pepper reference genes under different experimental conditions
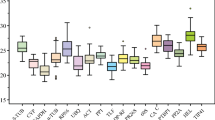
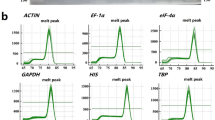
To select a set of reliable pepper reference genes for gene expression analysis, the qRT-PCR analysis method based on SYBR Green detection was used to evaluate the stability of expression of ten reference genes in different experimental conditions (Table 1). Melting curve analysis of amplification products confirmed that, at the expected melting temperature (T m), the primers amplified a single product (Electronic Supplementary Material). All the PCR assays were done in triplicate in order to confirm the reliability of the results. The Ct values of each reference gene in different experimental conditions were used to compare the various degrees of their expression levels. Figure 1 shows a relatively wide range of Ct values for the ten reference genes evaluated. The highest Ct value reached 34.36 (GAPDH), while the lowest was 8.16 (18S rRNA). Most of the remaining Ct values were distributed between 23 and 28. In addition, each individual reference gene had different Ct values in all of the applied conditions.
Evaluation of expression stability of putative reference genes
Expression profiles for each reference gene were analyzed using two different software methods, geNorm and NormFinder.
geNorm analysis
Expression stabilities (M values) of the ten candidates reference genes were calculated using the geNorm software and the reference genes were listed in descending order (most stable to least stable) of their stability according to their M values (Table 2). However, the ranking of the ten candidate reference genes was not consistent among individual pepper samples, as shown in the output from the geNorm analysis. The most stable reference gene varied according to the tissue used or experimental condition employed.
For cold- and heat-stress treatments, expression levels of the Actin1 and CYC genes were the most stable. Actin1 and GAPDH exhibited the most stable expression levels in all of the ten reference genes under salt- and drought-stress treatments, while EF1α and UEP were the most stable reference genes under the different hormone treatments and Actin and EF1α were the most stable genes in different plant tissues. Generally, our study revealed that Actin and Actin1 were the most stable reference genes in all the sample pools tested. They may therefore be useful in multiple reference experimental conditions. GAPDH was the least stable of the reference genes evaluated.
The results described above indicate that the expression levels of the ten genes varied according to the experimental conditions. For example, the CYC gene was one of the two most stable genes in cold- and heat-stress treatments, whereas it was the least stable in salt- and drought-stress treatments; this was also the case with the GAPDH gene between salt- and drought-stress treatments and different plant tissues. Therefore, it is necessary to choose one or more appropriate reference gene(s) before performing experiments to obtain reliable target gene results.
In order to determine the optimal number of reference genes in each experimental condition, pair-wise variation (V n /V n+1) was calculated by geNorm. The cut-off value usually used for determining the optimal number of reference genes is 0.15, below which the inclusion of additional reference genes is not required (Vandesompele et al. 2002). It should be noted that 0.15 is not an absolute cut-off value but an ideal value. Whether 0.15 is used as a cut-off value or not will depend on the data. Figure 2 shows the pair-wise variation values in each experimental set. Analysis of the pair-wise variation in cold- and heat-stress treatments revealed that the V 2/3 value was 0.009 (significantly <0.15), indicating that these two reference genes, Actin1 and CYC, were sufficient for normalization of gene expression. Similarly, in salt- and drought-stress treatments, Actin1 and GAPDH were the optimal normalization genes for gene expression analysis. In addition, evaluation of different hormones and different tissues experimental data sets showed that EF1α/UEP and Actin/EF1α were the top reference genes and are suitable for normalization of gene expression, respectively. However, evaluation of all the samples tested gave a V 3/4 value of 0.145, which was below the cut-off value of 0.15, indicating that three reference genes were sufficient for gene expression studies.
Pair-wise variation (V) analysis of the ten candidate reference genes. The pairwise variation (V n /V n+1) was analyzed between the normalization factors NF n and NF n+1 by geNorm software to determine the optimal number of reference genes required for qRT-PCR data normalization. Arrows indicate the best-suited pair-wise comparisons
NormFinder analysis
NormFinder is another algorithm for identifying the optimal normalization gene among a set of candidate reference genes (Andersen et al. 2004). It ranks the set of candidate normalization genes according to their expression stability in a given sample set and given experimental design. According to NormFinder, TEF1A was the most stable reference gene in cold- and heat-stress treatments. PPR1 exhibited the most stable expression levels in salt- and drought-stress treatments. For different hormone treatments and tissues, UEP was the most appropriate reference gene. Meanwhile, when all the results of the tested samples were jointly evaluated, UEP and PPR1 were the two most stable reference genes, followed by EF1α and Actin1 (Table 3).
The results obtained from NormFinder were not identical with those of geNorm except for hormone treatments, where the least stable gene was the same in every experimental set. However, GAPDH showed opposite results between geNorm and NormFinder in drought- and salt-stress treatments. The differences noted between the two methods were expected since the two programs are based on different statistical algorithms. geNorm relies on the principle that the expression ratio of two ideal reference genes is constant in all the samples, independent of the experimental conditions, while the algorithm of NormFinder uses a solid statistical framework to estimate not only the overall expression variation of the candidate reference genes but also the variation between subgroups of the sample sets. Notably, it provides a stability value for each gene, which is a direct measure of the estimated expression variation.
Discussion
qRT-PCR is currently one of the most widely used methods for gene expression analysis in different species (Piston et al. 2004; Doshi et al. 2007; Fang et al. 2009). However, it is essential to select stable reference genes for accurate normalization of gene expression. The ideal reference gene should have constant expression stability regardless of experimental conditions. However, no single gene has a stable expression under every experimental condition. Numerous studies have shown that the expression of even the housekeeping genes also varies considerably with different experimental conditions (Thellin et al. 1999). This may partly be explained by the fact that housekeeping genes are not only implicated in the basal cell metabolism but also participate in other cellular functions (Singh and Green 1993; Ishitani et al. 1996). In this present study, the feasibility of using ten putative reference genes as internal controls for gene expression studies in pepper was studied using geNorm and NormFinder, and the results showed that there were certain differences between the two programs with regard to identification of the best sets of reference genes for each experimental condition.
The data obtained showed that each experimental condition had a most suitable reference gene. We also demonstrated the significance of choosing suitable reference genes in gene expression studies for specific experimental conditions or different tissues. For example, EF1α and UEP were considered the most stable genes for gene expression normalization by both algorithms in different hormone treatments, but they were not the most stable genes in other experimental sets. However, the results were similar when all samples together were analyzed by both geNorm and NormFinder. Actin, Actin1, EF1α, and UEP were the most stable genes when calculated by geNorm. Whereas, when calculated by NormFinder, UEP, PPR1, EF1α, and Actin1 were the most stable genes. The calculation of V 3/4 by geNorm was 0.145, which was below the cut-off value of 0.15, indicating that three reference genes were sufficient for gene expression studies. Therefore, we recommend the use of EF1α, UEP, and Actin1 as suitable reference genes for normalization of gene expression in a wide range of samples tested in the present study. Although the use of the combination of EF1α, UEP, and Actin1 genes as an internal control may be sufficient for gene expression studies in pepper, a more accurate normalization can be performed if Actin is added, but the use of more than three control genes is unnecessary for the analyses (Erkens et al. 2006).
In conclusion, based on the results of the two methods, geNorm and NormFinder, EF1α, UEP, and Actin1 can be considered as suitable reference genes under all of the conditions tested in this study. These genes will enable more accurate and reliable normalization of qRT-PCR results for gene expression studies in pepper.
References
Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Bustin SA, Benes V, Nolan T, Pfaffl MW (2005) Quantitative realtime RT-PCR—a perspective. J Mol Endocrinol 34:597–601
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Dilger M, Felsenstein FG, Schwarz G (2003) Identification and quantitative expression analysis of genes that are differentially expressed during conidial germination in Pyrenophora teres. Mol Gen Genomics 270:147–155
Doshi KM, Eudes F, Laroche A, Gaudet D (2007) Anthocyanin expression in marker free transgenic wheat and triticale embryos. In Vitro Cell Dev Biol Plant 43:429–435
Erkens T, Van PM, Vandesompele J, Goossens K, Van ZA, Peelman LJ (2006) Development of a new set of reference genes for normalization of real-time RT-PCR data of porcine backfat and longissimus dorsi muscle, and evaluation with PPARGC1A. BMC Biotechnol 6:41
Fang CX, Xiong J, Qiu L, Wang HB, Song BQ, He HB, Lin RY, Lin WX (2009) Analysis of gene expressions associated with increased allelopathy in rice (Oryza sativa L.) induced by exogenous salicylic acid. Plant Growth Regul 57:163–172
Gachon C, Mingam A, Charrier B (2004) Real-time PCR: what relevance to plant studies? J Exp Bot 55:1445–1454
Ginzinger DG (2002) Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol 30:503–512
Giulietti A, Overbergh L, Valckx D, Decallonne B, Bouillon R, Mathieu C (2001) An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods 25:386–401
Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6(4):279–284
Ishitani R, Sunaga K, Hirano A, Saunders P, Katsube N, Chang DM (1996) Evidence that glyceraldehydes-3-phosphate dehydrogenase is involved in age-induced apoptosis in mature cerebellar neurons in culture. J Neurochem 66:928–935
Jarošova J, Kundu J (2010) Validation of reference genes as internal control for studying viral infections in cereals by quantitative real-time RT-PCR. BMC Plant Biol 10:146–154
Kim DH, Kang JG, Kim B (2007) Isolation and characterization of the cytoplasmic male sterility-associated orf456 gene of chill pepper (Capsicum annuum L.). Plant Mol Biol 63:519–532
Lland H, Hertzberg M, Marlton P (2006) Myeloid leukemia. In: Colgan SP (ed) Methods and protocols. Humana, Totowa, NJ, p 53
Narasimha RN, Harinath D, Yamini KN, Sujatha M, Dinesh KV (2009) Expression of sunflower cytoplasmic male sterility-associated open reading frame, orfH522 induces male sterility in transgenic tobacco plants. Planta 229:987–1001
Piston F, Dorado G, Martin A, Barro F (2004) Cloning and characterization of a gamma-3 hordein mRNA (cDNA) from Hordeum chilense. Theor Appl Genet 108:1359–1365
Remans T, Smeets K, Opdenakker K, Mathijsen D, Vangronsveld J, Cuypers A (2008) Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 227:1343–1349
Schmittgen TD, Zakrajsek BA (2000) Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods 46:69–81
Singh R, Green MR (1993) Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science 259:365–368
Sturzenbaum SR, Kille P (2001) Control genes in quantitative molecular biological techniques: the variability of invariance. Comp Biochem Physiol B Biochem Mol Biol 130:281–289
Thellin O, Zorzi W, Lakaye B, Borman DB, Coumans B, Hennen G, Grisar T, Igout A, Heinen E (1999) Housekeeping genes as internal standards: use and limits. J Biotechnol 75:291–295
Vandesompele J, De PK, Pattyn F, Poppe B, Van RN, De PA, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7): research0034.1-0034.11
Weis JH, Tan SS, Martin BK, Wittwe CT (1992) Detection of rare mRNAs via quantitative RT-PCR. Trends Genet 8:263–264
Yoo WG, Kim TI, Li S, Kwon OS, Cho PY, Kim T, Kim K, Hong S (2009) Reference genes for quantitative analysis on Clonorchis sinensis gene expression by real-time PCR. Parasitol Res 104:321–328
Yusuke I, Koji K, Kyonoshin M, Teruaki T, Masatomo K, Motoaki S, Kazuo S, Kazuo Y (2006) Functional analysis of rice DREB/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 47(1):141–153
Zhao HL, Ding LP, Sun JB, Wang SB (1995) Selection and identification of CMS lines 21A, 8A and 17A in pepper. Jiangsu Agric Sci 1:49–51
Acknowledgments
This research was supported by the General Program from the Natural Science Foundation of Jiangsu (BK2010464); National Staple Vegetables Industrial Technology System Huai’an Experiment Station Project; Jiangsu Programs CX (11)104 and CX (10)103.
Author information
Authors and Affiliations
Corresponding author
Additional information
Wang Shu Bin and Liu Ke Wei have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bin, W.S., Wei, L.K., Ping, D.W. et al. Evaluation of appropriate reference genes for gene expression studies in pepper by quantitative real-time PCR. Mol Breeding 30, 1393–1400 (2012). https://doi.org/10.1007/s11032-012-9726-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-012-9726-7