Abstract
The process of vernalization is mainly controlled by two genes in winter barley (Hordeum vulgare L.), VRNH1 and VRNH2. A recessive allele at VRNH1 and a dominant allele at VRNH2 must be present to induce a vernalization requirement. In addition, this process is usually associated with greater low-temperature tolerance. Spanish barleys originated in areas with mild winters and display a reduced vernalization requirement compared with standard winter cultivars. The objective of this study was to investigate the genetic origin of this reduced vernalization requirement and its effect on frost tolerance. We introgressed the regions of a typical Spanish barley line that carry VRNH1 and VRNH2 into a winter cultivar, Plaisant, using marker-assisted backcrossing. We present the results of a set of 12 lines introgressed with all four possible combinations of VRNH1 and VRNH2, which were evaluated for vernalization requirement and frost tolerance. The reduced vernalization requirement of the Spanish parent was confirmed, and was found to be due completely to the effect of the VRNH1 region. The backcross lines showed no decline in frost tolerance compared with that of the recurrent parent unless they carried an extra segment of chromosome 5H. This extra segment, a carryover of the backcross process, apparently contained the well-known frost tolerance quantitative trait locus Fr-H2. We demonstrate that it is possible to manipulate the vernalization requirement with only minor effects on frost tolerance. This finding opens the path to creating new types of barley cultivars that are better suited to specific environments, especially in a climate-change scenario.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The onset of flowering is one of the critical transitions in the life cycle of a plant, marked by the shift from the vegetative to the reproductive meristem stage. This process is regulated by a complex genetic system that combines internal developmental signals with environmental cues, such as temperature and photoperiod, and the interactions between them. This system must be tuned to guarantee that flowering takes place under suitable environmental conditions, so as to ensure optimal seed production (Izawa et al. 2003; Kim et al. 2009).
Vernalization accelerates heading by the promotion of inflorescence initiation of the shoot apex (Flood and Halloran 1984) and can be favoured by long-day conditions. The requirement of vernalization is especially involved in adaptation to winter temperatures (Trevaskis et al. 2003; Hemming et al. 2009).
Three loci, VRNH1, VRNH2 and VRNH3, control the vernalization requirement in barley (Hordeum vulgare L.) (Takahashi and Yasuda 1971; Distelfeld et al. 2009). The epistatic interaction between VRNH1 and VRNH2 is responsible for the presence of a winter growth habit (Takahashi and Yasuda 1971; Laurie et al. 1995; Yan et al. 2003, 2004; von Zitzewitz et al. 2005). The presence of a dominant VRNH2 allele is needed for a strict winter growth habit, although more recently obtained results suggest that this gene participates in photoperiod rather than low-temperature sensing (Distelfeld et al. 2009). VRNH1 apparently presents an allelic series that, together with a dominant VRNH2, determines the gradation of vernalization needs (Takahashi and Yasuda 1971; Szűcs et al. 2007).
Two major loci that control frost tolerance have been identified in temperate cereals (reviewed by Galiba et al. 2009): Frost Resistance 1 (Fr-1) and Frost Resistance 2 (Fr-2) (Hayes et al. 1993; Galiba et al. 1995; Francia et al. 2004). Both of these loci reside on chromosome 5 and are about 20–50 cM apart (Vágújfalvi et al. 2000; Tóth et al. 2003; Francia et al. 2004). In barley, Fr-H1 co-segregates with VRNH1, whereas Fr-H2 is in a 0.8-cM chromosomal region that encompasses a cluster of genes which encode C-repeat binding factors (Vágújfalvi et al. 2003; Skinner et al. 2006; Tondelli et al. 2006; Francia et al. 2007). Whether Fr-H1 and VRNH1 are two independent genes or the pleiotropic effects of the same gene is still unresolved.
Winter cereals, such as barley, are most productive when given a long growing season, that is, when they are sown in autumn. Most Spanish barley landraces have a reduced vernalization requirement compared with standard winter cultivars (Ciudad et al. submitted for publication); in other words, they have an intermediate vernalization requirement. Intermediate vernalization alleles might offer advantages in autumn-sowing areas like the Mediterranean region, where winters are not as cold as those in more northerly or continental climates, but still prevent the widespread use of spring cultivars. In the future, if global warming progresses as predicted, these alleles could become useful in regions currently dominated by strict winter cultivars.
Molecular tools have become integral to the enhancement of breeding efficiency and effectiveness in many research programmes. One of the main applications of molecular markers in breeding is to facilitate the targeted introgression of desirable traits into adapted cultivars through marker-assisted backcrossing, even for adaptation/productivity-related traits (Neeraja et al. 2007; Ribaut and Ragot 2007; Levi et al. 2009). In this study, we used marker-assisted backcrossing to introduce the regions that carry the two main vernalization genes, VRNH1 and VRNH2, from a Spanish landrace into a winter cultivar. Near-isogenic lines (NILs) introgressed with genomic regions that contained different vernalization loci (VRNH2 on chromosome 4H and VRNH1 on 5H) were developed and tested under controlled conditions. The objectives were to find out which of these regions is responsible for the reduced vernalization response of the Spanish landrace, and to measure their effects on vernalization requirement and frost tolerance.
Materials and methods
Plant material
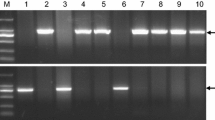
An introgression backcross program was carried out using marker-assisted selection, as reported by Frisch et al. (1999) and Frisch and Melchinger (2001). The recurrent parent was the French barley cultivar Plaisant (Ager × Nymphe), which is a typical winter cultivar that carries a dominant allele in VRNH2 and a recessive vrnh1 allele (full-length intron). The donor line was SBCC058, an inbred line derived from a Spanish landrace which belongs to the Spanish Barley Core Collection (Igartua et al. 1998). SBCC058 is an intermediate cultivar that carries the dominant allele in VRNH2 and a partial deletion in VRNH1, similar to Albacete or Calicuchima-sib (von Zitzewitz et al. 2005; Szűcs et al. 2007), which seems to be associated with a reduced vernalization requirement (Cockram et al. 2007; Hemming et al. 2009). VRNH1 was genotyped in both parents using markers already reported by von Zitzewitz et al. (2005), as shown in Electronic Supplementary Material Fig. S1. The NILs studied were derived from BC3F3 families. For the marker-assisted backcross (MAB) scheme, SBCC058/Plaisant F1s were backcrossed to Plaisant to obtain BC1F1 seeds. In the BC1F1 generation, individual plants that were heterozygous at the VRNH2 (flanking markers HvM067 and HdAMYB) and VRNH1 (flanking markers Hv635P2 and scssr10148) regions were identified (forward selection). The heterozygous plants were further screened with other markers to select plants that were homozygous for the recurrent genotype just outside both regions (backward elimination). A similar strategy was adopted in BC2F1 and BC3F1, namely, forward selection of heterozygous plants in the region of both VRN genes and backward elimination of the SBCC058 genome, with sequential addition of markers for the rest of the seven chromosomes (up to 79 simple sequence repeats [SSRs], Electronic Supplementary Material Table S1). HvBM5A, the candidate gene for VRNH1 (von Zitzewitz et al. 2005), was used from BC2F1 onwards. Homozygous plants for VRNH1 and VRNH2 were selected in BC3F2. On the basis of the allelic combination for these genes, four groups were established (Table 1). Group 1 referred to NILs that contained VRNH1 and VRNH2, such as Plaisant, and this group was used to check the effectiveness of recovery of the recurrent genotype. Group 2 was NILs that had VRNH1 from Plaisant and VRNH2 from SBCC058. Group 3 NILs had VRNH1 from SBCC058 and VRNH2 from Plaisant. Group 4 NILs carried VRNH1 and VRNH2 from SBCC058.
Vernalization treatments
The vernalization requirement was evaluated at the Martonvásár (Hungary) Phytotron, according the procedures described by Karsai et al. (2004). Vernalization was applied in 15-day increments, for a total of four treatments that ranged from no vernalization to 45 days of vernalization at a temperature below 3°C, with an 8-h light/16-h dark photoperiod and low light intensity (12–13 μmol m−2 s−1). After the vernalization treatment, seedlings were transferred to a 16 h light regime (340 ± 22 μmol m−2 s−1) at a constant temperature of 18°C. Two plants per genotype and treatment were tested. For each plant, the number of days to heading [namely, developmental phase 49 on the Zadoks scale (Tottman and Makepeace 1979)], number of tillers at heading, number of leaves on the main shoot, and number of reproductive tillers were recorded.
The frost tolerance of the BC3F3 plants, together with that of the two parents and check cultivars, was evaluated in artificial freezing tests in which two freezing temperatures of −11.5 or −13°C were applied in accordance with the methodology of Tischner et al. (1997). After germination, the seedlings were planted in wooden boxes and were acclimatized to cold for 7 weeks under conditions of weekly decreasing temperature, photoperiod, and light intensity. After gradual cooling, the frost tests were carried out in C-912 Conviron chambers (Conviron Ltd., Winnipeg, MB, Canada) for 24 h at the freezing temperature, which was followed by gradual warming. The plants were then grown under optimal conditions for 3 weeks for recovery, and the percentages of plants that survived were recorded. Five replications of ten plants per genotype and treatment were tested.
Field trials
The BC3F3 family NILs, the parents (SBCC058 and Plaisant), and several cultivars that were included as checks were tested in the field in the 2008–2009 season. The first trial was sown in autumn, on 27 November 2008, and the second one was sown in winter, on 12 February 2009. Both trials took place in Zaragoza (Spain). Plots were 1.5 m wide by 3 m (first sowing) or 1.2 m (second sowing) long, with four rows. A randomized complete block design with three replications was used. The heading date of each plot was recorded as the date when 50% of the stems presented 2-cm emerged awns. The time to flowering was expressed as the number of days from 1 January until heading time.
SNP genotyping
To estimate the recovery of the genome of Plaisant through the MAB process, the 12 BC3F3 NILs plus the parents were genotyped for 1,536 single nucleotide polymorphisms (SNPs) using Barley Oligo Pool Assay 1 (BOPA) of Illumina (Close et al. 2009) at the Southern California Genotyping Consortium. Genomic DNA was extracted from one individual plant of each genotype.
Statistical analyses
To evaluate the effect of the introgressed regions, differences among the four groups of NILs and the parents were evaluated together using ANOVA and orthogonal contrasts. In these ANOVA models, replication, genotype (NILs) and treatment were considered as fixed factors. Genotype by treatment interaction was also included in the models. The variability due to replicates and their interaction with the other factors was used as the error term to test genotype, treatment, and their interaction. All the analyses were carried out with GenStat 12.1 (VSN International Ltd., UK).
Results
Recovery of the recurrent parent
Out of the 1536 SNPs evaluated, 1313 had a known position in a genetic map and only 459 of these were polymorphic between the parents of this cross. The 12 NILs that were obtained covered 92% of the genome of Plaisant, with slight differences between them (Table 2). As expected, the highest recovery was for group 1, whose plants were similar to Plaisant in both 4H and 5H, and where recovery should have been 100%. The lowest percentages occurred for plants of group 4, with introgression of SBCC058 regions in both 4H and 5H. Graphical genotypes are shown in Fig. 1 (detailed information in Fig. S2).
Vernalization requirement
There were significant differences among the groups of NILs for time to flowering and other variables, in response to the duration of the vernalization treatment (Table S2). Most of the genotypic variation detected occurred between groups (97.5%), and this was mostly due to significant contrasts between groups 1/2 and 3/4, and between the parents (Table S2). The interaction of genotype with treatments presented a similar pattern, with most of the interaction being due to the different response of flowering time of groups 1 and 2 compared with that of groups 3 and 4 (Fig. 2a, Table S2) in response to the duration of the cold treatment. This means that the introgression of the SBCC058 region with VRNH1 reduced the vernalization requirement, whereas there was no difference between the SBCC058 and Plaisant VRNH2 regions. Groups 1 and 2, which carry the Plaisant allele in VRNH1, behaved in similar ways. In these groups, flowering did not occur without vernalization, and was still remarkably late after 15 days of cold treatment, and became earlier with each vernalization time increment. This pattern of response was similar to that of Plaisant. Given the trend observed for these groups and for the parent Plaisant, it is likely that longer vernalization would have further shortened the time to flowering. Groups 3 and 4, which share the SBCC058 VRNH1 allele, also presented similar performances. These lines flowered much earlier than groups 1 and 2 under all conditions (Fig. 2a), but still with a clear and significantly shorter interval after 15 days of vernalization. Parent SBCC058 responded to vernalization in a similar way to groups 3 and 4, although it flowered later than these groups in all treatments. A similar result was detected for the number of reproductive tillers at heading, with a clear difference between groups and their differential response to vernalization (Fig. 2b, Table S2). Owing to the fact that all the groups consistently flowered only after 30 and 45 days of cold treatment, we only analysed the differences for number of tillers at heading and number of leaves on the main shoot for these two treatments (Fig. 2c, d, Table S2), as it was not possible to get realistic estimates of them for the treatments of 0 and 15 days. Again, plants that carried the introgression of the SBCC058 VRNH1 allele were more stable in their behaviour, and exhibited fewer tillers at heading and fewer leaves than those with the Plaisant VRNH1 allele.
Days to flowering (a), number of reproductive tillers (b), number of tillers at heading (c), and number of leaves on the main shoot (d) of BC3F3 NILs of the cross between the donor genotype SBCC058 and the recurrent parent Plaisant, after 0, 15, 30, or 45 days of vernalization. Error bars correspond to least significant differences of means (1% level). Genotype codes: 1a to 4d, NILs; 58, SBCC058; PL, Plaisant; DT, Dicktoo; MX, Morex
Frost tolerance
Almost all the NILs showed a good survival rate, around 90% at −11.5°C and around 70% at −13°C, close to the values of the parent Plaisant (Fig. 3a, b), and most of them were more frost-tolerant than the donor parent SBCC058. There were significant differences between genotypes for both treatments (Table S3), but these were not concentrated among groups. At −11.5°C, the only significant differences detected were the higher tolerance of groups 3 and 4 compared with SBCC058. At −13°C, genotypic differences were again not related to comparisons between the groups (Table S3). This means that, in general, the introgressed regions did not affect frost tolerance. The genotypic differences were caused by a markedly lower frost tolerance observed for some NILs. NIL 4b at both temperatures, and NIL 4c at −13°C, showed significantly reduced frost tolerance compared with the other NILs (even in the same group), and reached values close to those of SBCC058 (Fig. 3b). Examination of the graphical genotypes (Fig. 1 and Fig. S2) revealed that these two plants shared some regions with residual alleles of SBCC058 at chromosomes 2H, 4H and 5H, which were outside the target regions of the introgression program. The factor behind their lower frost tolerance must be located in these regions. The most likely candidate is the region on 5H just above VRNH1, which carries the well-known frost resistance locus Fr-H2.
To test whether VRNH1 itself had an effect on freezing tolerance, independent of other regions that may affect frost tolerance, we carried out new analyses of variance without NILs that carried the Fr-H2 region from SBCC058 (1b, 4b and 4c). Significant differences between groups were now detected, but concentrated in the differences of the recurrent parent SBCC058 from all the introgression groups and Plaisant (Fig. 3, Table S3). Actually, none of the introgression groups was significantly different from the recurrent parent Plaisant. The contrast between VRNH1 alleles (groups 1 and 2 vs. groups 3 and 4) was barely significant (P = 0.012).
Field flowering date
Significant differences in time to flowering in the field were found between genotypes and groups. In the group sown in autumn, all the introgression lines flowered around the same date as Plaisant. Only SBCC058 flowered significantly later (Fig. 4). The differences between groups and genotypes were larger in the group sown in late winter (Fig. 4), for which groups 1 and 2 and Plaisant were significantly later than the rest. This behaviour resulted in the occurrence of significant sowing by genotype interactions, which was caused mainly by the differences in response between sowing for groups 1 and 2 and Plaisant versus groups 3 and 4 and SBCC058. As described above for the Phytotron experiment, the differences were based on the VRNH1 region, whereas no significant differences between the two VRNH2 alleles were detected (Table S4).
Discussion
The MAB program led to the efficient integration of VRNH1 and VRNH2 from the Spanish landrace SBCC058 into the background of the winter cultivar Plaisant, with a good recovery of the recurrent genome (average 95.9%). Overall, the recovery of the recurrent genome was good in all chromosomes not directly affected by the introgression. It was better for chromosomes 1H, 2H, 3H and 7H, whereas for 6H recovery was only 93.1%, which indicated that the tagging of this chromosome during the process was not optimal. The values for chromosomes 4H and 5H, on which the VRN genes are located, were slightly lower (91.8%). In particular, the region distal to VRNH1 on 5H was more diverse than the others. Selection for the VRNH1 gene was carried out using markers within the VRNH1 candidate gene HvBM5A, and using the microsatellite Bmag0223 as a flanking marker. These two markers are 50 cM apart in the map of the Beka × Mogador population (Cuesta-Marcos et al. 2008) or 30 cM apart according to a new barley integrated map (Aghnoum et al. 2010). In either case, the gap between these markers proved to be too large to facilitate clean recovery of the recurrent parent.
Plants were grouped on the basis of genetic constitution for VRNH1 and VRNH2 into four groups made by plants from independent lineages. Only two plants from group 4 (4b and 4c) were sister lines. Both parents contained the VRNH2 gene, although flanking markers differed between them, which facilitated tracking of the two parental segments during the introgression. The comparison of the two VRNH2 alleles did not reveal any phenotypic difference in any of the experiments; both of them seemed equally functional. The objective of this work was not to establish sequence differences between the parents, but the data available confirms that we have two winter alleles. A partial sequence (1470 bp) of the HvZCCT-Hb gene from SBCC058 was 100% identical to that of Calicuchima-sib (GenBank accession DQ492696), and slightly different from the sequences of Oregon Wolfe Barley Dominant, Dairokkaku, and Kompolti korai (Szűcs et al. 2007), all of them carrying functional winter VRNH2 alleles. We did not sequence the allele from Plaisant, but it presumably carries a typical winter VRNH2 allele. The same cannot be said for VRNH1. The allele from SBCC058 clearly reduced the vernalization requirement of Plaisant and enabled plants to flower earlier in the Phytotron and in the field under late winter sowing. This effect was detected irrespective of the VRNH2 allele present.
Plants introgressed with the SBCC058 VRNH1 allele showed a small but clear response to vernalization in the Phytotron experiment: on average, they flowered 22 days earlier when fully vernalized. This behaviour was significantly different from the responses of the facultative cultivar Dicktoo (5 days of cycle shortening due to vernalization) and of the typical spring cultivar Morex (0 days). However, their behaviour was also clearly different from the typical winter behaviour, with a strict vernalization requirement, here represented by Plaisant and groups 1 and 2. Takahashi and Yasuda (1971) have described a continuous gradation of growth habit in barley, from an extreme spring to an extreme winter habit. They classified plants into six growth habit classes and concluded that a multiple allelic series at the VRNH1 locus is responsible for the grade of growth habit. Hemming et al. (2009) have identified 10 alleles that contain insertions or deletions within the first intron of VRNH1, and are associated with different flowering behaviour. In addition to the wild-type winter allele VRNH1 in cultivars Plaisant or Dicktoo, two other alleles were involved in our experiments: VRNH1-1 in cultivar Morex and VRNH1-4 in the breeding line Calicuchima-sib. Both of these deletions have been described previously (von Zitzewitz et al. 2005; Cockram et al. 2007; Szűcs et al. 2007), and were found to be associated with a reduced vernalization requirement. SBCC058 carries the same alleles at VRNH1 (VRNH1-4) and VRNH2 as Spanish cultivars Albacete and Pané or the breeding line Calicuchima-sib (von Zitzewitz et al. 2005; Szűcs et al. 2007). To date, these kinds of genotypes with the VRNH1 allele of SBCC058 and VRNH2 have been classified agronomically as spring varieties that show a reduced vernalization requirement (Cockram et al. 2007; Szűcs et al. 2007; Hemming et al. 2009). This allelic constitution is representative of a large class of Spanish barleys that have been classified as exhibiting a winter growth habit on the basis of their suitability to be sown in autumn and their mild vernalization requirement (Lasa et al. 2001). As proposed by Szűcs et al. (2007) for Calicuchima-sib, SBCC058 represents an intermediate growth habit, which corresponds to one of the intermediate classes of Takahashi and Yasuda (1971).
The donor line, SBCC058, showed a larger vernalization response (39 days) than NILs with the same VRNH1-4 allele in a Plaisant background. This genotype also flowered later than Plaisant and all the introgression lines sown in autumn in the field, which indicated that other genes besides VRNH1 and VRNH2 could have affected the flowering time of SBCC058.
Genetic analyses have identified two loci in wheat and barley that mediate the capacity to overwinter in temperate climates. One locus (Fr-1) co-segregates with VRNH1, which affects the vernalization requirement. The latest data from wheat indicate that Fr-1 is a pleiotropic effect of VRN1 rather than a separate gene (Dhillon et al. 2010). The second locus, Fr-2, is coincident with a cluster of more than 12 CBF genes (Francia et al. 2004; Skinner et al. 2006; Galiba et al. 2009), which are also located on chromosome 5H.
We found frost tolerance differences between the NILs that belonged to group 4, particularly after exposure to a temperature of −13°C. It has been reported that the VRNH1/Fr-H1 locus affects the expression of multiple barley CBF genes at Fr-H2 (Stockinger et al. 2007). However, the donor genotype SBCC058 and two lines of group 4 were extremely susceptible to frost, but other lines with the same SBCC058 VRNH1 allele behaved similarly to the winter recurrent parent. Dhillon et al. (2010) showed that allelic variation in wheat VRN1 is sufficient to determine differences in freezing tolerance, but additional genes are needed for down-regulation of the cold acclimation pathway. We hypothesize that differences between lines 4b and 4c and the other lines were mainly due to Fr-H2, because they were polymorphic for SNPs from that region on chromosome 5H. VRNH1 is located at 135.7 cM on 5H, as indicated by the barley BOPA consensus map of Close et al. (2009). In 4b and 4c, the SBCC058 introgressed region extended further, from 107.59 to 137.16 cM. Plants 4b and 4c differed in four SNP markers, between 11_20549 at 108.01 cM and 11_21168 at 109.56 cM, because 4b was homozygous for the SBCC058 allele, whereas 4c was heterozygous (Fig. S2). This is the region that corresponds to the cluster of cold-related CBF genes, at 108.2 cM, and indeed one of the SNPs scored, 11_20320, was derived from HvCBF6.
One of the lines of group 1, NIL 1b, also carried the Fr-H2 region introgressed from SBCC058. However, the frost tolerance of this line was not significantly lower than that of the other lines of groups 1 and 2, or that of Plaisant. The reason for this could be that it carries the Plaisant VRNH1 strict winter allele. This would conform to the hypothesis of interaction between vernalization requirement and frost tolerance put forward by Galiba et al. (2009) and Dhillon et al. (2010), that the temperate cereals have evolved the ability to use the presence of VRN1 in the leaves as a signal to down-regulate the frost/cold tolerance genes, such as Fr-H2. Akar et al. (2009) have reported that HvBM5A, the marker derived from the candidate genes VRNH1 and Fr-H1, is the best predictor for assisted selection to improve frost tolerance in European germplasm, with a larger effect than markers at the Fr-H2 region. We detected a possible effect of VRNH1 on frost tolerance, after removal of the two NILs of group 4 with reduced tolerance, but the effect was not large enough to reach definitive conclusions.
Our results demonstrate that the SBCC058 VRNH1 allele produced important changes in the developmental pattern of the recurrent parent Plaisant, whereas VRNH2 had no effect (possibly, the allele was the same for both parents). In addition, VRNH1 from SBCC058, typical of a large group of Spanish barleys, cannot be described as a spring or a winter allele. Its behaviour fits well with an intermediate position along the gradient of growth habits described by Takahashi and Yasuda (1971). An important finding for barley breeding is that it is possible to manipulate the vernalization requirement, with little effect on frost tolerance. This result widens the prospects of managing adaptation genes to breed new cultivars that are better suited to a range of winter harshness.
References
Aghnoum R, Marcel TC, Johrde A, Pecchioni N, Schweizer P, Niks RE (2010) Basal host resistance of barley to powdery mildew: connecting quantitative trait loci and candidate genes. Mol Plant Microbe Interact 23:91–102
Akar T, Francia E, Tondelli A, Rizza F, Stanca AM, Pecchioni N (2009) Marker-assisted characterization of frost tolerance in barley (Hordeum vulgare L.). Plant Breed 128:381–386
Close TJ, Bhat PR, Lonardi S, Wu Y, Rostoks N, Ramsay L, Druka A, Stein N, Svensson JT, Wanamaker S, Bozdag S, Roose ML, Moscou MJ, Chao S, Varshney RK, Szűcs P, Sato K, Hayes PM, Matthews DE, Kleinhofs A, Muehlbauer GJ, DeYoung J, Marshall DF, Madishetty K, Fenton RD, Condamine P, Graner A, Waugh R (2009) Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics 10:582
Cockram J, Chiapparino E, Taylor SA, Stamati K, Donini P, Laurie DA, O’Sullivan DM (2007) Haplotype analysis of vernalization loci in European barley germplasm reveals novel VRN-H1 alleles and a predominant winter VRN-H1/VRN-H2 multi-locus haplotype. Theor Appl Genet 115:993–1001
Cuesta-Marcos A, Igartua E, Ciudad FJ, Codesal P, Russell JR, Molina-Cano JL, Moralejo MA, Szűcs P, Gracia MP, Lasa JM, Casas AM (2008) Heading date QTL in a spring × winter barley cross evaluated in Mediterranean environments. Mol Breed 21:455–471
Dhillon T, Pearce SP, Stockinger EJ, Distelfeld A, Li C, Knox AK, Vashegyi I, Vágújfalvi A, Galiba G, Dubcovsky J (2010) Regulation of freezing tolerance and flowering in temperate cereals: the VRN-1 connection. Plant Physiol. doi:10.1104/pp.110.159079
Distelfeld A, Li C, Dubcovsky J (2009) Regulation of flowering in temperate cereals. Curr Opin Plant Biol 12:178–184
Flood RG, Halloran GM (1984) The nature and duration of gene action for vernalization response in wheat. Ann Bot 53:363–368
Francia E, Rizza F, Cattivelli L, Stanca AM, Galiba G, Toth B, Hayes PM, Skinner JS, Pecchioni N (2004) Two loci on chromosome 5H determine low-temperature tolerance in a ‘Nure’ (winter) × ‘Tremois’ (spring) barley map. Theor Appl Genet 108:670–680
Francia E, Barabaschi D, Tondelli A, Laidò G, Rizza F, Stanca AM, Busconi M, Fogher C, Stockinger EJ, Pecchioni N (2007) Fine mapping of a HvCBF gene cluster at the frost resistance locus Fr-H2 in barley. Theor Appl Genet 115:1083–1091
Frisch M, Melchinger AE (2001) Marker-assisted backcrossing for simultaneous introgression of two genes. Crop Sci 41:1716–1725
Frisch M, Bohn M, Melchinger AE (1999) Comparison of selection strategies for marker-assisted backcrossing of a gene. Crop Sci 39:1295–1301
Galiba G, Quarrie SA, Sutka J, Morgaunov A, Snape JW (1995) RFLP mapping of the vernalization (Vrn1) and frost resistance (Fr1) genes on chromosome 5A of wheat. Theor Appl Genet 90:1174–1179
Galiba G, Vágújfalvi A, Li C, Solté A, Dubcovsky J (2009) Regulatory genes involved in the determination of frost tolerance in temperate cereals. Plant Sci 176:12–19
Hayes PM, Blake T, Chen THH, Tragoonrung S, Chen F, Pan A, Liu B (1993) Quantitative trait loci on barley (Hordeum vulgare L.) chromosome-7 associated with components of winter hardiness. Genome 36:66–71
Hemming MN, Fieg S, Peacock WJ, Dennis ES, Trevaskis B (2009) Regions associated with repression of the barley (Hordeum vulgare) VERNALIZATION1 gene are not required for cold induction. Mol Genet Genomics 282:107–117
Igartua E, Gracia MP, Lasa JM, Medina B, Molina-Cano JL, Montoya JL, Romagosa I (1998) The Spanish barley core collection. Genet Resour Crop Evol 45:475–481
Izawa T, Takahashi Y, Yano M (2003) Comparative biology comes into bloom: genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Curr Opin Plant Biol 6:113–120
Karsai I, Hayes PM, Kling J, Matus IA, Mészáros K, Láng L, Bedő Z, Sato K (2004) Genetic variation in component traits of heading date in Hordeum vulgare subsp. spontaneum accessions characterized in controlled environments. Crop Sci 44:1622–1632
Kim DH, Doyle MR, Sung S, Amasino RM (2009) Vernalization: winter and the timing of flowering in plants. Annu Rev Cell Dev Biol 25:277–299
Lasa JM, Igartua E, Ciudad FJ, Codesal P, Garcia EV, Gracia MP, Medina B, Romagosa I, Molina-Cano JL, Montoya JL (2001) Morphological and agronomical diversity patterns in the Spanish barley core collection. Hereditas 135:217–225
Laurie DA, Pratchett N, Bezant JH, Snape JW (1995) RFLP mapping of five major genes and eight quantitative trait loci controlling flowering time in a winter × spring barley (Hordeum vulgare L) cross. Genome 38:575–585
Levi A, Paterson AH, Barak V, Yakir D, Wang B, Chee PW, Saranga Y (2009) Field evaluation of cotton near-isogenic lines introgressed with QTLs for productivity and drought related traits. Mol Breed 23:179–195
Neeraja CN, Maghirang-Rodriguez R, Pamplona A, Heuer S, Collard BCY, Septiningsih EM, Vergara G, Sanchez D, Xu K, Ismail AM, Mackill DJ (2007) A marker-assisted backcross approach for developing submergence-tolerant rice cultivars. Theor Appl Genet 115:767–776
Ribaut JM, Ragot M (2007) Marker-assisted selection to improve drought adaptation in maize: the backcross approach, perspectives, limitations, and alternatives. J Exp Bot 58:351–360
Skinner JS, Szűcs P, von Zitzewitz J, Marquez-Cedillo L, Filichkin T, Stockinger EJ, Thomashow MF, Chen TH, Hayes PM (2006) Mapping of barley homologs to genes that regulate low temperature tolerance in Arabidopsis. Theor Appl Genet 112:832–842
Stockinger EJ, Skinner JS, Gardner KG, Francia E, Pecchioni N (2007) Expression levels of barley Cbf genes at the Frost resistance-H2 locus are dependent upon alleles at Fr-H1 and Fr-H2. Plant J 51:308–321
Szűcs P, Skinner JS, Karsai I, Cuesta-Marcos A, Haggard KG, Corey AE, Chen THH, Hayes PM (2007) Validation of the VRN-H2/VRN-H1 epistatic model in barley reveals that intron length variation in VRN-H1 may account for a continuum of vernalization sensitivity. Mol Genet Genomics 277:249–261
Takahashi R, Yasuda S (1971) Genetics of earliness and growth habit in barley. In: Nilan RA (ed) Barley genetics II. Washington State University Press, Washington, pp 388–408
Tischner T, Kőszegi B, Veisz O (1997) Climatic programmes used in the Martonvásár phytotron most frequently in recent years. Acta Agron Hung 45:85–104
Tondelli A, Francia E, Barabaschi D, Aprile A, Skinner JS, Stockinger EJ, Stanca AM, Pecchioni N (2006) Mapping regulatory genes as candidates for cold and drought stress tolerance in barley. Theor Appl Genet 112:445–454
Tóth B, Galiba G, Fehér E, Sutka J, Snape JW (2003) Mapping genes affecting flowering time and frost resistance on chromosome 5B of wheat. Theor Appl Genet 107:509–514
Tottman DR, Makepeace RJ (1979) An explanation of the decimal code for the growth stages of cereals, with illustrations. Ann Appl Biol 93:221–234
Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES (2003) MADS box genes control vernalization-induced flowering in cereals. Proc Natl Acad Sci USA 100:13099–13104
Vágújfalvi A, Galiba G, Cattivelli L, Dubcovsky J (2003) The cold regulated transcriptional activator Cbf3 is linked to the frost-tolerance locus Fr-A2 on wheat chromosome 5A. Mol Genet Genomics 269:60–67
Vágújfalvi A, Crosatti C, Galiba G, Dubcovsky J, Cattivelli L (2000) Two loci on wheat chromosome 5A regulate the differential cold-dependent expression of the cor14b gene in frost-tolerant and frost-sensitive genotypes. Mol Gen Genet 263:194–200
von Zitzewitz J, Szűcs P, Dubcovsky J, Yan L, Pecchioni N, Francia E, Casas A, Chen THH, Hayes PM, Skinner JS (2005) Molecular and structural characterization of barley vernalization genes. Plant Mol Biol 59:449–467
Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100:6263–6268
Yan L, Loukoianov A, Blech A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303:1640–1644
Acknowledgments
This work was funded by the Spanish Ministry of Science and Innovation (projects AGL2004-05311, GEN2006-28560-E, and AGL2007-63625), and co-funded by the European Regional Development Fund. CC was supported by an I3P predoctoral fellowship from CSIC.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11032_2010_9497_MOESM2_ESM.doc
Supplementary material 2 Fig. S1 PCR amplification of the region harbouring the 5.2 kb intron 1 InDel using primer set HvBM5A.055F/056R (von Zitzewitz et al. 2005). SBCC058 and Plaisant are depicted.(DOC 42 kb)
11032_2010_9497_MOESM3_ESM.xls
Supplementary material 3 Fig. S2 BOPA1 genotypes of BC3F3 NILs of the cross SBCC058 × Plaisant. 1 (green, Plaisant allele); 2 (red, SBCC058 allele); 3 (orange, heterozygous SNP).(XLS 127 kb)
Rights and permissions
About this article
Cite this article
Casao, M.C., Igartua, E., Karsai, I. et al. Introgression of an intermediate VRNH1 allele in barley (Hordeum vulgare L.) leads to reduced vernalization requirement without affecting freezing tolerance. Mol Breeding 28, 475–484 (2011). https://doi.org/10.1007/s11032-010-9497-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-010-9497-y