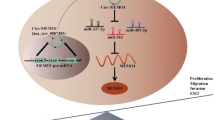
Abstract
Colorectal cancer (CRC) remains a malignancy tumor with high metastasis and poor prognosis. We aimed to explore the effect of circular RNA (circRNA) hsa_circ_0006732 in the progression of CRC. Hsa_circ_0006732 expression in CRC tissues and cell lines were detected using qRT-PCR. The relationship between hsa_circ_0006732 expression and clinicopathologic characteristics of patients with CRC was analyzed. Loss-of-function assay was conducted to determine the regulatory effect of hsa_circ_0006732 on CRC cell proliferation, migration and invasion by using the CCK-8, wound-healing assay and transwell assays. Protein expression changes on epithelial mesenchymal transition (EMT)-related factors were detected by western blotting. The downstream signaling pathway was investigated by bioinformatics, dual-luciferase reporter assay. Rescue assay was further examined for prediction validation. It was found that hsa_circ_0006732 was highly expressed in CRC tissues and cell lines. Downregulation of hsa_circ_0006732 suppressed the proliferation, migration, invasion and EMT of CRC cells. Further mechanistic investigations proved that hsa_circ_0006732 functioned as a competitive endogenous RNA (ceRNA) by directly sponging of miR-127-3p, which further affected the expression of Ras-related protein Rab-3D (Rab3D). Taken together, these findings indicated that hsa_circ_0006732 might be an oncogene in CRC through the regulation of the miR-127-5p/RAB3D axis. Thus, hsa_circ_0006732 might serve as a potential therapeutic target for the treatment of CRC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) refers to the malignant tumor of colon and rectum [1]. According to the survey from the International Center for research on cancer, there were 1.8 million new cases of CRC in 2018, and about 880,000 people lost their lives due to CRC and its complications [2]. The mortality and incidence of CRC cancer ranked second and third among all malignant tumors, respectively. In 2019, the incidence rate of newly diagnosed CRC cases in China was 376 thousands (8%), and about 191 thousands (7%) died of CRC, ranking fifth in morbidity and mortality [3]. Many evidences show that high fat diet promotes the occurrence of CRC. Part of the food is decomposed by the flora in the digestive tract, and the products change the microecological environment of intestinal epithelial cells, and the microecological environment also affect the growth of intestinal flora [4]. The two interact with each other to participate in the occurrence and development of CRC [5]. Genetic factors play an important role in the occurrence of CRC. Studies have shown that oncogene and tumor suppressor gene mutation, DNA sequence mutation, DNA methylation and histone acetylation are involved in the development of CRC [6]. However, the exact pathogenesis of CRC is still unknown.
Circular RNA (circRNA) is characterized by single stranded circular structure [7]. Due to its special closed loop structure, circRNA is not easy to be degraded by RNase [8]. Some studies have shown that circRNA is involved in a variety of biological processes, such as competitive binding of endogenous RNA, chelating proteins, regulation of gene transcription and translation [9]. CircRNAs affect initial immune response, neural function, cell proliferation and stem cell pluripotency [10]. Some circRNAs are involved in the development of diabetes, atherosclerosis and other diseases [10]. In addition to benign diseases, circRNAs is closely related to malignant tumors and plays a role in promoting or inhibiting the development of various malignant tumors.
At present, a large number of circRNAs have not been deeply studied, and some studies have shown that circRNAs may be involved in the occurrence and development of CRC, such as hsa_circ_0004585 [11], hsa_ circ_0026344 [12] and hsa_circ_0005273 [13]. The study of circRNAs can help people understand the pathogenesis of CRC and provide research basis for the diagnosis and treatment of CRC. According to the CircBase database, hsa_circ_0006732, also known as circRNA zinc finger DHHC-type palmitoyltransferase 20 (circZDHHC20), was located at chr13:21987790–21999817. Hsa_circ_0006732 was reported to be a tumor suppressor in preeclampsia, and it suppressed trophoblast cells migration, proliferation and invasion in preeclampsia through regulating miR-144 to modulate GRHL2 [14]. Up to now, the expression and role of hsa_circ_0006732 in CRC have not been reported.
In the present study, we found that hsa_circ_0006732 was highly expressed in CRC tumor tissues and cell lines and related to the prognosis of CRC patients. And, we tested the role of hsa_ circ_ 0006732 played in the occurrence and development of CRC, and further explored the function of hsa_circ_0006732 in CRC cells. In this study, a preliminary study on the diagnosis and treatment of hsa_circ_0006732 in CRC was conducted, in order to provide a research basis for the diagnosis and treatment of CRC.
Materials and methods
Patient tissues
Human CRC tissues (n = 108) and matched paracancerous normal intestinal tissues (n = 108) were acquired from CRC patients from the First Affiliated Hospital of Jinzhou Medical University Hospital during operation. Liquid nitrogen was used to store these above tissues in a refrigerator at − 80 °C. This study was approved by the Ethics Committee of the First Affiliated Hospital of Jinzhou Medical University Hospital. The written informed consent was signed by the relevant patients in our study.
Cell culture
Human CRC cell lines (T84, LoVo, SW480, HCT116, HCT8), and normal colonic epithelial cells (NCM460) were all purchased from the Procell Life Science & Technology (Wuhan, China). DMEM, provided by Invitrogen (Carlsbad, CA, USA), was used to culture the above cell lines with fetal bovine serum (FBS, 10%, Invitrogen), streptomycin (100 μg/ml, Invitrogen) and penicillin (100 U/ml) at 37 °C in 5% CO2.
Cell transfection
Two small interfering (si) RNAs (si-circRNA#1 and si-circRNA#2) that against hsa_circ_0006732 and their negative control (si-NC), miR-127-5p inhibitor (miR-127-5p inh), and the over-expression plasmid of RAB3D (oe-RAB3D) were all synthesized by VectorBuilder (Guangzhou, China). The cell transfection was proceeded using Lipofectamine 2000 (Thermo Fisher, USA) according to the manufacturer's protocol.
Cell counting kit-8 (CCK-8) assay
96-well plates were employed to seed the cells used in our study with a denstiy of 2 × 103 cells/well. CCK-8 solution that bought from Dojindo (10 μL, Tokyo, Japan) was supplemented into each well and incubated for 2 h. A microplate reader was applied to observe and record the optical density (OD) values at 0, 24, 48 and 72 h after transfection.
Quantitative real time polymerase chain reaction (qRT-PCR)
A trizol reagent, purchased from Beyotime (Shanghai, China), was used to lyse the CRC cells and tissues. A NanoDrop-1000 apparatus that provided by Thermo Fisher was used to measure the RNA concentration. A PrimeScript RT Master Mix, provided by Takara (Dalian, China), was used to amplify cDNA. For determining the expression of hsa_circ_0006732, miR-127-5p, RAB3D, SYBR Green SuperMix, harvested from Roche (Basel, Switzerland) was performed. The 2−∆∆Ct method was used to analyze data. GAPDH or U6 were selected as references. The primers in this part were hsa_circ_0006732 forward: 5′-CCTACATTGACATGTACACAGAACA-3′, reverse: 5′-TTCCACTGATCATTTTCTTGC-3′; RAB3D forward: 5′-GCTGGTGGAAGATGGTTC-3′, RAB3D reverse: 5′-AAAGGCTGCTTCAATGCT-3′; miR-127-5p forward: 5′-GCTGAAGCTCAGAGGGC-3′, reverse: 5′-GTTGTGGTTGGTTGGTTTGT-3′; RAB3D forward: 5′-GCTGGTGGAAGATGGTTC-3′, reverse: 5′-AAAGGCTGCTTCAATGCT-3′; U6forward: 5′-ATTGGAACGATACAGAGAAGATT-3′, reverse: 5′-GGAACGCTTCACGAATTTG-3′; GAPDH forward: 5′-TCGGAGTCAACGGATTTGGT-3′, reverse: 5′-TTCCCGTTCTCAGCCTTGAC-3′.
Wound-healing assay
6-well culture plates were used to seed the cells at a density of 5 × 105 cells/well. The monolayer was scratched with new pipette tips (200 μL) across the well center after 24 h incubation. After scratching, the wells were washed twice and then replenished with fresh medium without FBS. After 24 h, closure of the gap was estimated by taking photographs. The software-ImageJ was carried out to quantitatively evaluate the distance.
Transwell assay
Millicell cell culture inserts (8-lm pore size, 24-well insert) were employed to conduct transwell assay. Matrigel, provided by BD Biosciences (NJ, USA), was coated to inserts, and then transfected cells that seeded in serum-free medium were added into the upper chamber (8 × 104 cells/well). Medium including FBS (10%) was added into the lower chamber. After incubation for 24 h, crystal violet solution (0.5%) was used to fix and stain the cell lines on the bottom of the inserts.
Western blot
Total protein was extracted by RIPA Lysis Buffer, bought from Sangon Biotech (Shanghai, China), was used to extract the total protein. And, a BCA Protein Assay Kit that was provided by Sangon Biotech was used to quantify the total protein. SDS-PAGE gel (10%) and PVDF membrane (Beyotime) were then respectively used to isolate and transfer the proteins. Nonfat milk (5%) were used to immerse the membranes, and then primary antibodies were added. The primary antibodies used in our study including anti-E-cadherin (ab40772, 1:40,000, Abcam, MA, USA), anti-Vimentin (ab92547, 1:1000), anti-MMP-2 (ab92536, 1:1000, Abcam), anti-MMP-9 (ab76003, 1:1000), anti-RAB3D (ab128997, 1:10,000), anti-GAPDH (ab9485, 1:2500). Subsequently, Goat Anti-Rabbit IgG H&L (HRP) (ab205718, 1:50,000) was added. After that, an ECL luminescent solution, harvested from Meilunbio (Dalian, China), was used to analyze the protein signals.
Bioinformatics analysis
Circbase (http://www.circbase.org) was used to obtain the hsa_circ_0006732 sequence. Starbase (http://starbase.sysu.edu.cn/mirCircRNA.php), Circular RNA Interactome (https://circinteractome.nia.nih.gov), and TargetScan (http://www.targetscan.org/) used to predict the binding sites of hsa_circ_0006732, miRNAs, and the target mRNAs of miR-127-5p.
Dual-luciferase reporter assay
pmirGLO vector, provided by Promega, was used to clone the hsa_circ_0006732 or RAB3D 3'UTR with binding sites for mutant (MUT) or wild-type (WT) (WT-circ_0006732, MUT-circ_0006732, WT-RAB3D or MUT-RAB3D). MiR-127-5p mimic (or miR-NC), and the corresponding luciferase reporter were co-transfected into the CRC cells. A dual-luciferase reporter kit (Beyotime) was employed to measure the luciferase activity.
Statistical analysis
Mean ± standard deviation (SD) was used to express all statistical data in three independent experiments via SPSS 19.0. Student’s T-test or one-way analysis of variance (ANOVA) followed by Tukey’s test were both used to assess the differences. p < 0.05 was considered as statistically significant. Correlation analysis was conducted using Pearson coefficient correlation.
Results
Hsa_circ_0006732 was up-regulated in CRC tissues and cell lines
We detected hsa_circ_0006732 expression levels in 108 pairs of CRC tumor tissues hsa_circ_0006732 expression in normal group was significantly lower than that in tumor group (p < 0.01, Fig. 1A). Then, we found that hsa_circ_0006732 expression was associated with CRC clinical-pathological features, including tumor size and TNM stage (p < 0.05, Table 1). In addition, data from qRT-PCR displayed that the hsa_circ_0006732 expression in human colon epithelial NCM460 cells was remarkably lower than that in CRC tumor cell lines including T84, LoVo, SW480, HCT116, HCT8 (p < 0.05, Fig. 1B). And, CRC cell lines HCT116 and HCT8 were chosen to carry out following experiments due to the higher hsa_circ_0006732 expressions. These results displayed that hsa_circ_0006732 was upregulated in CRC tumor tissues and cell lines.
Hsa_circ_0006732 was up-regulated in CRC tissues and cell lines. A The expression level of hsa_circ_0006732 in 108 pairs of CRC tumor tissues and normal tissues was detected using qRT-PCR. B The expression level of hsa_circ_0006732 in CRC cell lines was detected using qRT-PCR. Data were shown as mean ± SD. **p < 0.01 vs Normal group or NCM460 cells
Hsa_circ_0006732 knockdown inhibited cell biological activities in CRC cell lines
To downregulate the hsa_circ_0006732 expression in HCT116 and HCT8 cells, si-circRNA#1 and si-circRNA#2 were synthesized and transfected into HCT116 and HCT8 cells. As shown as Fig. 2A, both si-circRNAs transfection significantly decreased hsa_circ_0006732 expression in CRC cells (p < 0.01), and si-circRNA#1 showed the better knockdown efficiency. Therefore, si-circRNA#1 was selected to conduct next experiments. Data from CCK-8 confirmed that the knockdown of hsa_circ_0006732 markedly reduced cell proliferation in both HCT116 and HCT8 cells (p < 0.01, Fig. 2B). And, we found that the si-circRNA group was significantly lower than that of si-NC group (p< 0.01, Fig. 2C). Then, wound healing assay was carried out to assess the cell migration, and the results of wound healing assay was showed as Fig. 2D. Obviously, the silencing of hsa_circ_0006732 suppressed the cell migration in both HCT116 and HCT8 cell lines (p < 0.01). Furthermore, the expressions of epithelial mesenchymal transition (EMT)-related proteins E-cadherin, Vimentin, MMP-2 and MMP-9, were detected using western blot. Data from western blot presented that the knockdown of hsa_circ_0006732 remarkably decreased the Vimentin, MMP-2 and MMP-9 expressions and increased the E-cadherin expression in both HCT116 and HCT8 cells (p < 0.01, Fig. 2E). These results confirmed that the knockdown of hsa_circ_0006732 inhibited cell proliferation, invasion, migration and EMT in CRC cell lines.
Knockdown of hsa_circ_0006732 inhibited cell biological activities in CRC cell lines. A The expression of hsa_circ_0006732 was detected using qRT-PCR. B Cell proliferation was assessed using CCK-8 assay. C Transwell assays were performed to detect changes in the invasion abilities of HCT116 and HCT8 cells transfected with si-hsa_circ_0006732 or si-NC. D Wound healing assay was used to determine the migratory abilities of HCT116 and HCT8 cells following transfection of si-hsa_circ_0006732 or si-NC. E The expressions of EMT-related proteins including E-cadherin, Vimentin, MMP-2 and MMP-9 were detected using western blot. Data were shown as mean ± SD. **p < 0.01 vs si-NC group
Hsa_circ_0006732 directly targeted to miR-127-5p
We predicted that hsa_circ_0006732 targeted to miR-127-5p by Circinteractome database and Starbase database, and the binding sites of hsa_circ_0006732 on miR-127-5p were showed in Fig. 3A. As shown as Fig. 3B, the WT-circ_0006732 and MUT-circ_0006732 vectors were constructed, and miR-127-5p mimic significantly reduced the relative luciferase activity of WT-circ_0006732 (p < 0.01) but did not change the activity of MUT-circ_0006732. Data from qRT-PCR displayed that the expression of miR-127-5p in normal tissues were markedly higher than that in CRC tumor tissues (p < 0.01, Fig. 3C). And, in CRC tumor tissues, the expression of hsa_circ_0006732 was negatively associated with the expression of miR-127-5p (Fig. 3D). In addition, the expression of miR-127-5p was remarkably enhanced with the knockdown of hsa_circ_0006732 in both HCT116 and HCT8 cell lines (p < 0.01, Fig. 3E). These data demonstrated that hsa_circ_0006732 sponged miR-127-5p in CRC.
Hsa_circ_0006732 directly targeted to miR-127-5p. A Circinteractome database and Starbase database were performed to predict the binding sites of hsa_circ_0006732 on miR-127-5p. B Luciferase reporter assay was carried out to verify the prediction. C The expression of miR-127-5p in 108 pairs of CRC tumor tissues and normal tissues was detected using qRT-PCR. D The relationship between hsa_circ_0006732 expression and miR-127-5p expression was analyzed using Pearson coefficient correlation. E The expression of miR-127-5p was detected using qRT-PCR. Data were shown as mean ± SD. **P < 0.01 vs si-NC group
MiR-127-5p directly targeted to RAB3D
We predicted that RAB3d was the downstream mRNA of miR-27-5p via Targetscan, and the binding sites of RAB3D on miR-127-5p were showed in Fig. 4A. As shown as Fig. 4B, the WT-RAB3D and MUT-RAB3D vectors were established, and miR-127-5p mimic significantly decreased the relative luciferase activity of WT-RAB3D (p < 0.01) but did not change the activity of MUT-RAB3D. Results from qRT-PCR displayed that the expression of RAB3D in CRC tumor tissues were markedly higher than that in normal tissues (p < 0.01, Fig. 4C). And, in CRC tumor tissues, the expression of RAB3D was positively associated with the expression of hsa_circ_0006732, and negatively related to the expression of miR-127-5p (Fig. 4D). In addition, the expression of RAB3D was remarkably reduced with the knockdown of hsa_circ_0006732 in both HCT116 and HCT8 cell lines (p < 0.01, Fig. 4E). These results suggested that RAB3D was targeted by miR-127-5p in CRC.
MiR-127-5p directly targeted to RAB3D. A Targetscan was performed to predict the binding sites of RAB3D on miR-127-5p. B Luciferase reporter assay was carried out to verify the prediction. C The expression of RAB3D in 108 pairs of CRC tumor tissues and normal tissues was detected using qRT-PCR. The relationship among RAB3D expression, D hsa_circ_0006732 expression and E miR-127-5p expression was analyzed using Pearson coefficient correlation. F The relative expression of RAB3D was detected using western blot using GAPDH as an internal control. Protein levels are shown as bar graphs. Data were shown as mean ± SD. **p < 0.01 vs si-NC group
Hsa_circ_0006732 regulated cell biological activities in CRC cell lines through miR-127-5p/RAB3D axis
To verify whether hsa_circ_0006732 acted on CRC by miR-127-5p/RAB3D axis, a rescue experiment was carried out. The HCT116 cells were divided into four groups including si-NC group, si-circRNA group, si-circRNA + miR-127-5p inh group and si-circRNA + oe-RAB3D group according to different treatments. CCK-8 assay showed that the effect of hsa_circ_0006732 knockdown on CRC cell proliferation was significantly recovered by the inhibition of miR-127-5p or the up-regualtion of RAB3D (p < 0.01, Fig. 5A). As shown as Fig. 5B andC, si-circRNA + miR-127-5p inh transfection or si-circRNA + oe-RAB3D transfection also eliminated the suppressive effect of hsa_circ_0006732 silencing on the invasion and migration of HCT116 cells (p < 0.01). In addition, the decreases of Vimentin, MMP-2 and MMP-9 expression and the increase of E-cadherin induced by si-circRNA transfection in CRC cells were reversed by the si-circRNA + miR-127-5p inh transfection or si-circRNA + oe-RAB3D transfection (p < 0.01, Fig. 5D). Therefore, we elucidated that hsa_circ_0006732 regulated cell proliferation, invasion, migration and EMT in CRC through miR-127-5p/RAB3D axis.
Hsa_circ_0006732 regulated cell biological activities in CRC cell lines through miR-127-5p/RAB3D axis. A Cell proliferation was assessed using CCK-8 assay. B Cell invasion was analyzed using transwell assay. C Cell migration was examined using wound healing assay. D The expressions of EMT-related proteins including E-cadherin, Vimentin, MMP-2 and MMP-9 were detected using western blot. Data were shown as mean ± SD. **p < 0.01 vs si-NC group, ##p < 0.01 vs si-circRNA group
Discussion
The occurrence of CRC is generally considered to be formed by a series of mutations in the epithelial cells of colon gland. In 1990, Feraron and Vogelstein first proposed that [15] tubular villous adenoma is a precancerous lesion that can develop into CRC. The process from precancerous lesions to cancer takes a long time, from several years to decades. With the improvement of science and technology, people realize that the types of precancerous lesions may be gradually increasing [16]. Previous studies found that the changes of various signal pathways can cause CRC. These changes of molecular signals are helpful to people to understand the development of CRC [17]. With the development of molecular pathway of CRC, precise treatment of CRC has been widely concerned. Many molecular targeted drugs such as bevacizumab, cetuximab, panimab and regofinib have been gradually applied in clinical practice [18]. The adverse reactions and limitations of these drugs in different situations force us to find more and effective treatment ways to improve the diagnosis and treatment level of CRC.
CircRNAs play a regulatory role in the development and progression of tumors. Several circRNAs were found to affect CRC in previous studies [19]. Wang et al. [20] reported that circ_0060745 enhanced CRC cells proliferation and metastasis that induced by chromosome segregation 1-like (CSE1L) and played as a competing endogenous RNA (ceRNA) to sequester miR-4736. Tang et al. [21] found that CircRNA circ_0124554 facilitated the skip lymphovascular invasion with hepatic metastasis, interdicted the ubiquitination of AKT in CRC. In the current study, we found that hsa_circ_0006732 was upregulated in CRC tumor tissues and cell lines. Then, the expression of hsa_circ_0006732 was knocked down in HCT116 and HCT8 cell lines by si-circRNA transfection. We confirmed that the cell proliferaion, migration and invasion were all inhibited with hsa_circ_0006732 silencing.
EMT is closely related to the invasion and metastasis of tumor, which attracts more and more attention [22]. The occurrence of EMT is accompanied by changes of several factors, such as downregulation of E-cadherin expression, upregulation of Vimentin, MMP-2 and MMP-9 [23]. In the present study, we found that the downregulation of hsa_circ_0006732 improved the expression of E-cadherin in CRC cells as well as repressed the expressions of Vimentin, MMP-2 and MMP-9 by western blot. It suggested that the knockdown of hsa_circ_0006732 blocked EMT in CRC cells.
CircRNAs were found to regulate downstream mRNA by acting as microRNA (miRNA) sponges. In our paper, we respectively predicted and verified that hsa_circ_0006732 sponged miR-127-5p in CRC cells through Circinteractome database and luciferase reporter assay. MiR-127-5p was found to impair tumor cell activity and function in various cancers, such as hepatocellular carcinoma [24], esophageal squamous cell carcinoma [25] and cervical cancer [26]. We demonstrated that the miR-127-5p was lowly expressed in CRC tissues. In CRC cell lines, we found that the expression of miR-127-5p was upregulated when the expression of hsa_circ_0006732 was downregulated. In CRC tumor tissues, the expression of miR-127-5p was negatively associated with the expression of hsa_circ_0006732.
In addition, we forecast that miR-127-5p targeted to RAB3D via Targetscan, and then validated the prediction by luciferase reporter assay. In previous studies, RAB3D was seemed as a oncogene in several cancers, such as non-small cell lung cancer [27], glioma [28], osteosarcoma [29] and CRC [30]. Then, we proved that the expression of RAB3D was positively associated with the expression of hsa_circ_0006732 in CRC tumor tissues, and negatively related to the expression of miR-127-5p. In HCT116 and HCT8 cells, we found that the expression of RAB3D was remarkably reduced with the knockdown of hsa_circ_0006732. Furthermore, a rescue experiment was carried out. We found that the inhibitions of cell proliferation, migration, invasion and EMT in CRC cells that induced by the knockdown of hsa_circ_0006732 were recovered by the inhibition of miR-127-5p or the up-regulation of RAB3D. However, several limitations were included in our study. First, further validating investigations may have to be performed using a larger cohort to provide a greater statistical significance. Second, the further investigation is needed to verify the in vivo role of hsa_circ_0006732 in CRC using xenograft model. Moreover, other possible mechanisms underlying hsa_circ_0006732 in CRC development were not deeply studied.
In conclusion, we found that hsa_circ_0006732 was highly expressed in CRC tumor tissues and cell lines, and indicated poor prognosis of CRC patients. And, hsa_circ_0006732 regulated CRC cell proliferation, invasion, migration and EMT through sponging miR-127-5p to control RAB3D. Hsa_circ_0006732 might be a potential therapeutic method against CRC.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
References
Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ (2017) Colorectal carcinoma: a general overview and future perspectives in colorectal cancer. Int J Mol Sci 18:197
Cronin KA, Lake AJ, Scott S, Sherman RL, Noone AM, Howlader N, Henley SJ, Anderson RN, Firth AU, Ma J (2018) Annual report to the nation on the status of cancer, part I: National cancer statistics. Cancer 124:2785–2800
Chen H, Li N, Ren J, Feng X, Lyu Z, Wei L, Li X, Guo L, Zheng Z, Zou S (2019) Participation and yield of a population-based colorectal cancer screening programme in China. Gut 68:1450–1457
Xie Y-H, Chen Y-X, Fang J-Y (2020) Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct Target Ther 5:1–30
Siegel RL, Fedewa SA, Anderson WF, Miller KD, Ma J, Rosenberg PS, Jemal A (2017) Colorectal cancer incidence patterns in the United States. JNCI. https://doi.org/10.1093/jnci/djw322
Thanikachalam K, Khan G (2019) Colorectal cancer and nutrition. Nutrients 11:164. https://doi.org/10.3390/nu11010164
Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu Y-M, Dhanasekaran SM, Engelke CG, Cao X (2019) The landscape of circular RNA in cancer. Cell 176(869–881):e13
Han B, Chao J, Yao H (2018) Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther 187:31–44
Zeng X, Lin W, Guo M, Zou Q (2017) A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput Biol 13:e1005420
Hsiao K-Y, Sun HS, Tsai S-J (2017) Circular RNA–new member of noncoding RNA with novel functions. Exp Biol Med 242:1136–1141
Tian J, Xi X, Wang J, Yu J, Huang Q, Ma R, Zhang X, Li H, Wang L (2019) CircRNA hsa_circ_0004585 as a potential biomarker for colorectal cancer. Cancer Manag Res 11:5413
Yuan Y, Liu W, Zhang Y, Zhang Y, Sun S (2018) CircRNA circ_0026344 as a prognostic biomarker suppresses colorectal cancer progression via microRNA-21 and microRNA-31. Biochem Biophys Res Commun 503:870–875
Yang H, Li X, Meng Q, Sun H, Wu S, Hu W, Liu G, Li X, Yang Y, Chen R (2020) CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol Cancer 19:1–15
Zhou B, Zhang X, Li T, Xie R, Zhou J, Luo Y, Yang C (2020) CircZDHHC20 represses the proliferation, migration and invasion in trophoblast cells by miR-144/GRHL2 axis. Cancer Cell Int 20:1–11
Fearon ER, Vogelstein B (1990) A genetic model for colorectal tumorigenesis. Cell 61:759–767
Weinberg BA, Marshall JL, Salem ME (2017) The growing challenge of young adults with colorectal cancer. Oncology (Williston Park) 31:381–389
Rawla P, Sunkara T, Barsouk A (2019) Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Przeglad Gastroenterologiczny 14:89
Grasso CS, Giannakis M, Wells DK, Hamada T, Mu XJ, Quist M, Nowak JA, Nishihara R, Qian ZR, Inamura K (2018) Genetic mechanisms of immune evasion in colorectal cancer. Cancer Discov 8:730–749
Lei B, Tian Z, Fan W, Ni B (2019) Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci 16:292–301
Wang X, Ren Y, Ma S, Wang S (2020) Circular RNA 0060745, a novel circRNA, promotes colorectal cancer cell proliferation and metastasis through miR-4736 sponging. Oncol Targets Ther 13:1941
Tang J, Zhang C, Huang Y, Wang L, Xu Z, Zhang D, Zhang Y, Peng W, Feng Y, Sun Y (2021) CircRNA circ_0124554 blocked the ubiquitination of AKT promoting the skip lymph invasion on hepatic metastasis in colorectal cancer. Cell Death Dis 12:1–12
Yang J, Antin P, Berx G, Blanpain C, Brabletz T, Bronner M, Campbell K, Cano A, Casanova J, Christofori G (2020) Guidelines and definitions for research on epithelial–mesenchymal transition. Nat Rev Mol Cell Biol 21:341–352
Dongre A, Weinberg RA (2019) New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol 20:69–84
Zhang W, Zhu L, Yang G, Zhou B, Wang J, Qu X, Yan Z, Qian S, Liu R (2020) Hsa_circ_0026134 expression promoted TRIM25-and IGF2BP3-mediated hepatocellular carcinoma cell proliferation and invasion via sponging miR-127-5p. Biosci Rep. https://doi.org/10.1042/BSR20191418
Xiong G, Diao D, Lu D, Liu X, Liu Z, Mai S, Feng S, Dong X, Cai K (2020) Circular RNA circNELL2 Acts as the sponge of miR-127-5p to promote esophageal squamous cell carcinoma progression. Onco Targets Ther 13:9245
Chang S, Sun L, Feng G (2019) SP1-mediated long noncoding RNA POU3F3 accelerates the cervical cancer through miR-127-5p/FOXD1. Biomed Pharmacother 117:109133
Li L, Wan K, Xiong L, Liang S, Tou F, Guo S (2020) CircRNA hsa_circ_0087862 acts as an oncogene in non-small cell lung cancer by targeting miR-1253/RAB3D axis. Onco Targets Ther 13:2873
Jin T, Liu M, Liu Y, Li Y, Xu Z, He H, Liu J, Zhang Y, Ke Y (2020) Lcn2-derived circular RNA (hsa_circ_0088732) inhibits cell apoptosis and promotes EMT in glioma via the miR-661/RAB3D Axis. Front Oncol 10:170
Jiashi W, Chuang Q, Zhenjun Z, Guangbin W, Bin L, Ming H (2018) MicroRNA-506-3p inhibits osteosarcoma cell prolif and metastasis by suppressing RAB3D expression. Aging (Albany NY) 10:1294
Luo Y, Yu S-Y, Chen J-J, Qin J, Qiu Y-E, Zhong M, Chen M (2018) MiR-27b directly targets Rab3D to inhibit the malignant phenotype in colorectal cancer. Oncotarget 9:3830
Acknowledgements
Not applicable.
Funding
The authors declare that they have no funding support.
Author information
Authors and Affiliations
Contributions
TY, JFS conceived and designed the study. WW and DSL performed the literature search and data extraction. XXY, AJ, YDM and ZF drafted the manuscript. All authors had read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
The experimental protocol was established, according to the ethical guidelines of the Helsinki Declaration and was approved by the Animal Ethics Committee of First Affiliated Hospital of Jinzhou Medical University.
Consent for publication
The authors agree to publication in the Journal.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yang, T., Sun, J., Wang, W. et al. Hsa_circ_0006732 regulates colorectal cancer cell proliferation, invasion and EMT by miR-127-5p/RAB3D axis. Mol Cell Biochem 477, 2751–2760 (2022). https://doi.org/10.1007/s11010-022-04458-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-022-04458-5