Abstract
Circular RNAs (circRNAs) play regulatory roles in the biological processes of multiple tumors, colorectal cancer (CRC) included. Our previous study probed the impact of circ_0007385 on CRC cell malignant behaviors, while the underlying mechanism remains obscure. In this work, the potential mechanism of hsa_circ_0007385 in CRC was probed. Functional experiments were implemented for probing the function of hsa_circ_0007385 in CRC. Further analysis revealed the relation between hsa_circ_0007385 and miRNAs. A xenograft mouse model was implemented for probing the influence of hsa_circ_0007385 on CRC growth and metastasis in vivo. Hsa_circ_0007385 was up-regulated in CRC. Hsa_circ_0007385 positively regulated its host gene mediator of cell motility 1 (MEMO1). Hsa_circ_0007385 silencing inhibited CRC progression. Hsa_circ_0007385 and MEMO1 bond to miR-485-3p/miR-543/miR-337-3p, and these three miRNAs were lowly expressed in CRC, and negatively modulated by hsa_circ_0007385. Hsa_circ_0007385 functioned as an oncogene in CRC in a miR-485-3p/miR-543/miR-337-3p- or MEMO1-dependent manner. Hsa_circ_0007385 promoted CRC progression via modulating miR-485-3p/miR-543/miR-337-3p/MEMO1 axis. Thus, circ-MEMO1 might be a promising therapeutic target for CRC.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is one of the most frequent cancers all over the world (Dekker et al. 2019). The incidence and mortality rate have been rising in recent years due to the changes in dietary structures and living habits (Bin et al. 2022). Late diagnosis is associated with the reduced survival in CRC patients (Saucedo-Sariñana et al. 2022). However, in spite of the improvements in the diagnosis together with therapy of CRC, the 5-year survival in CRC patients is 64.7% and their prognosis is still unsatisfactory (Chen et al. 2021a; Li and Mohammadi 2023). The exploration of molecular biomarkers with high sensitivity and specificity is proposed as an effective and reliable method for timely diagnosis in cancer patients (Tourang 2021). Current treatment options for CRC include surgery, radiotherapy, chemotherapy, immunotherapy, and targeted therapy (Alavi 2022; Biller and Schrag 2021; Tabin et al. 2022; Bilal 2021; Xu and Yang 2022). In clinic, metastasis is regarded as the major cause of death in CRC patients (Li et al. 2020).Distant tumor metastasis and recurrence remains a main obstacle for CRC treatment (Liu et al. 2022), and the long-term survival time of metastatic CRC patients is below 20% (Biller and Schrag 2021). Therefore, it is urgent and necessary to understand the underlying mechanism in CRC and develop effective therapeutic strategies.
Noncoding RNAs are critically involved in the initiation and progression of diverse cancers and provide promising therapeutic targets for the anti-cancer treatment (Yan and Bu 2021; Alsaedy et al. 2022). As a novel class of noncoding RNA that extensively expressed in eukaryotes, circular RNA (circRNA) is featured with a covalently closed ring structure (Zhu et al. 2021). Former reports consider that circRNAs are a by-product or the outcome of mis-splicing during pre-mRNA processing, and possess no biological properties (Zhou et al. 2020). With the advancement of RNA sequencing technology, the crucial biological properties as well as the therapeutic potential of circRNA is increasingly revealed (Kristensen et al. 2022), and the sponge-like potential circRNAs in cancer progression has attracted growing attention (Chen et al. 2021b). MicroRNAs (miRNAs) are small noncoding transcripts with 19–25 nucleotides in length and can interact with and regulate the expression of targeted mRNAs by promoting degradation or repressing translation (Azizi Dargahlou et al. 2023). Studies have also revealed the biological roles of miRNAs as oncogenes or cancer suppressor in the regulation of cellular processes in various cancer progression, and many miRNAs are suggested as diagnostic biomarkers or therapeutic targets (Kanwal et al. 2023; Chen et al. 2022; He and Zhou 2022). Many mRNAs and circRNAs share binding sequences with miRNAs in the regulation of cancer development, and mRNAs or circRNAs will compete for interaction with miRNAs, which is called competing endogenous RNA pattern (ceRNA) (Yang et al. 2022). Aberrant expression of circRNA is observed in the development of multiple human tumors (Liu et al. 2019). For instance, circRNA_0000285 is highly expressed in cervical cancer samples and accelerates cervical cancer cell growth, migration, and invasion ability as well as the tumorigenesis by up-regulating FUS (Chen et al. 2019). CircFOXK2 is up-regulated in breast cancer and its overexpression promotes cancer cell migration and invasiveness by interacting with IGF2BP3 or miR-370 (Zhang et al. 2021a). CircRNA_0000392 is expressed at higher levels in CRC tissues and its deficiency suppresses the proliferation and invasiveness of CRC cells in vitro and in vivo by acting as a ceRNA sponging miR-193a-5p to regulate PIK3R3 and affect AKT signaling activation (Xu et al. 2020a). Besides, numerous studies have revealed that circRNA-based diagnostic or therapeutic methods exhibit significant potential in cancer management (Li et al. 2021). Nevertheless, unlike microRNAs (miRNAs), the function of most circRNAs in modulating cancer progression is largely obscure. In view of the crucial roles of circRNAs in tumorigenesis, the investigation of circRNAs is essential.
In our previous study, we probed the influence of circ_0007385 on the CRC progression. This work aimed to further elucidate the biological roles and underlying regulatory mechanism of circ_0007385 in the CRC progression in vitro and in vivo. We hypothesized that circ_0007385 might affect CRC cell malignant behaviors and tumor growth by acting as a ceRNA to regulate the downstream gene expression, which might offer novel therapeutic targets for CRC treatment.
Materials and Methods
Tissue Samples
The application of tissue samples was authorized by the Ethics Committee of The First Affiliated Hospital of Soochow University. Thirty cases of CRC tumor samples together with adjacent normal tissues were obtained from The First Affiliated Hospital of Soochow University. Informed consent was acquired from all patients included in this study.
RT-qPCR
Total RNA was extracted using Trizol reagent (Invitrogen), and subsequently reverse-transcribed into cDNA by Prime Script RT Master Mix (Takara, Japan). RT-qPCR was conducted using GoTaq qRT-PCR Master Mix (Promega, USA). Gene expression was calculated based on 2−ΔΔCt method. GAPDH together with U6 served as internal references. The primer sequences of RNAs were presented below: Circ-MEMO1: F: 5′-ATGTTTGAACGCATGTCTCTGC-3′; R: 5′-CCGGGTAATAGACGGATCCAC-3′. MEMO1: F: 5′-GATTTACGGAGAACTGTGGA-3′, R: 5′-TCCTTATGGCTTTCCATGG-3′. MiR-485-3p: F: 5′-GCCGAGGUCAUACACGGCUCU-3′, R: 5′-CTCAACTGGTGTCGTGGA-3′. MiR-543: F: 5′-CAGTGCTAAAACATTCGCGG-3′, R: 5′-TATGGTTGTTCACGACTCCTTCAC-3′. MiR-337-3p: F: 5′-CUCCUAUAUGAUGCCUUUCUUC-3′, R: 5′-GAAGAAAGGCAUCAUCUAGGAG-3′. GAPDH: F: 5′-TCAAGATCATCAGCAATGCC-3′, R: 5′-CGATACCAAAGTTGTCATGGA-3′.
Cell Culture and Treatment
ATCC supplied CRC cell lines (SW480, LoVo, HCT-116, as well as Caco-2) together with human normal colonic epithelial cell line (HCoEpiC). SW480 together with HCoEpiC cells were cultured in Dulbecco’s Modified Eagle medium. LoVo cells were incubated in Ham’s F-12K medium. HCT-116 cells were cultured in McCoy’s 5A medium. Caco-2 cells were incubated in MEM medium. All medium was supplemented with 1% antibiotics together with 10% FBS and maintained at 37 °C with 5% CO2. Besides, cells were incubated with RNase R (Epicentre, USA) for 10 min. Then, circ-MEMO1 and MEMO1 expression was measured using RT-qPCR.
Cell Transfection
Two shRNAs targeting circ-MEMO1 and negative control, pcDNA-circ-MEMO1 and empty pcDNA, pcDNA-MEMO1 and empty pcDNA, miR-483-5p mimic, miR-543 mimic, miR-337-3p mimic together with NC mimic, as well as miR-483-5p inhibitor, miR-543 inhibitor, miR-337-3p inhibitor together with NC inhibitor were commercially acquired from GenePharma (Shanghai, China). Cell transfection was implemented using Lipofectamine™ 3000 (Life Technologies, USA).
Colony Formation Assay
Cells were planted into 12-well plates. Cells were immobilized by 4% paraformaldehyde and taken for staining using 1% crystal violet solution after incubation for 10 days. Cell colonies were counted and imaged.
TUNEL Assay
Cells were immobilized by 4% paraformaldehyde, and permeabilized in 100% methanol, followed by treatment with fluorescein-TUNEL reagent as well as DAPI. Apoptotic cells were visualized using an inverted microscope.
Transwell Assay
Transfected cells were put into the upper chamber of Transwell chambers (Corning, USA) for cell migration test. Cells were planted in the upper chambers pre-coated with Matrigel (BD Biosciences) for cell invasion test. Medium with 10% FBS in the lower chambers served as a chemoattractant. Cells above the surface were wiped off. Cells in the lower chamber were immobilized, stained, and counted under a light microscope.
Fluorescence In Situ Hybridization (FISH)
Circ-MEMO1 probe was labeled using Cy3 by Biosearch Technologies. For FISH assay, cells were grown in 24-well plates. After immobilization as well as permeabilization, cells were hybridized with Cy3-labeled circ-MEMO1 probe, and DAPI was used for cell nuclei staining. The images were captured by a florescent microscope.
Immunofluorescence (IF)
CRC cells were immobilized using 4% paraformaldehyde, followed by permeabilization using 0.1% Triton X-100 in PBS, and sealing with 5% BSA. Next, cells were incubated with anti-E-cadherin as well as anti-N-cadherin primary antibody at 4 °C overnight. After washing, cells were treated with corresponding secondary antibody and staining with DAPI. Images were captured by a confocal microscope.
Western Blot
Cells were lysed in radioimmunoprecipitation assay buffer (Sigma, USA). Protein extract were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, shifted to a polyvinylidene fluoride membrane (Millipore, USA) and blocked using skimmed milk. The membrane was cultured with anti-MEMO1 at 4 °C overnight, followed by treatment with secondary antibody (Abcam) and then visualized.
RNA-Binding Protein Immunoprecipitation (RIP)
This experiment was implemented using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA) following the producer’s instructions. Cells were treated with RIP lysis buffer, and added with anti-Ago2 or anti-IgG. Co-precipitated RNAs were purified, and assessed by RT-qPCR.
Dual-Luciferase Reporter Gene Assay
MiR-485-3p/miR-543/miR-337-3p and MEMO1 3’UTR wild-type (Wt) or mutant-type (Mut) were cloned into the luciferase reporter vector pmirGLO (Promega). CRC cells were transfected with the above luciferase plasmids together with pcDNA-circ-MEMO1, miR-485-3p mimic, miR-543 mimic, and miR-337-3p mimic. Forty-eight hours later, the luciferase activity was tested by the Dual-Luciferase Reporter Assay System (Promega).
Construction of shRNA Lentiviral Vectors
To repress the generation of circ-MEMO1, circ-MEMO1 shRNA or scrambled shRNA was constructed into BLOCK-iT™ Lentiviral RNAi expression system (Invitrogen) in LoVo cells. RT-qPCR was implemented to validate the transfection efficiency.
In Vivo Mouse Model
NOD. CB17-Prkdcscid/J (NOD SCID) mice were acquired from The First Affiliated Hospital of Soochow University, and all procedures acquired permission from the Animal Care and Use Committee. LV-sh-circ-MEMO1 transfected LoVo cells (1 × 106) or LV-sh-NC transfected LoVo cells were subcutaneously injected into the mice. The mice were euthanized after 27 days, and the transplanted tumor tissue was harvested. The tumor was excised and its weight and volume was determined.
Hematoxylin and Eosin (H&E) Staining
Mice tumor tissues were treated with 10% formaldehyde solution, dehydrated in an ethanol gradient, embedded in paraffin, as well as cut into sections in 4 μm thickness. Then samples were stained using hematoxylin together with eosin after deparaffinization, followed by observation under microscope.
Statistical Analysis
All experiments were carried out for three times. Statistical analyses were analyzed based on GraphPad Prism 7 together with SPSS 20.0. Student’s t test or one-way analysis of variance (ANOVA) was implemented for comparing the differences between groups. P < 0.05 was statistically significant.
Results
Hsa_circ_0007385 Positively Regulates MEMO1 Expression in CRC Cells
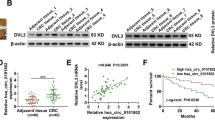
As indicated in violin plots and wire scatter plots, circ_0007385 was highly expressed in CRC tumor tissues (Fig. 1A, B). Consistently, RT-qPCR outcomes showed that circ_0007385 expression was elevated in 4 types of CRC cell lines, and was most significantly up-regulated in LoVo together with HCT-116 cells (Fig. 1C). Therefore, both LoVo and HCT-116 cells were selected for subsequent experiments. Based on UCSC (https://genome.ucsc.edu/) website, circ_0007385 was observed to be derived from the exon 3, 4, 5 loop of MEMO1 pre-mRNA, so circ_0007385 could also be called circ-MEMO1. To certify the circular properties of circ-MEMO1, we treated CRC cells with RNase R, and discovered that circ-MEMO1 expression had no change following RNase R addition, whereas MEMO1 mRNA was significantly reduced, indicating that circ_0007385 was stable (Fig. 1D). We designed 2 shRNAs targeting circ-MEMO1 to silence circ-MEMO1 expression, and adopted RT-qPCR analysis to certify the transfection efficiency. As displayed in Fig. 1E, circ-MEMO1 expression was successfully declined in CRC cells after transfection. As reported previously, MEMO1 is up-regulated in colon cancer cells (Xu et al. 2020b), thus we speculated that circ-MEMO1 might positively regulate MEMO1 expression in CRC cells. As expected, after circ-MEMO1 silence, MEMO1 expression was decreased in CRC cells (Fig. 1F). Additionally, ENCORI (http://starbase.sysu.edu.cn/) also exhibited that MEMO1 expression was increased in colon adenocarcinoma (COAD), a type of CRC (Fig. 1G). Moreover, MEMO1 was also up-regulated in CRC tumor tissues (Fig. 1H, I) and CRC cells (Fig. 1J). To further confirm the positive relation between circ-MEMO1 and MEMO1, FISH-IF experiment was implemented for examining the subcellular localization of circ-MEMO1 and MEMO1 in CRC cells. It was discovered that circ-MEMO1 and MEMO1 were mainly co-presented in the cytoplasm of CRC cells (Fig. 1K). All these outcomes indicated that circ-MEMO1 positively regulated MEMO1 expression in CRC.
Hsa_circ_0007385 positively regulates MEMO1 expression in CRC cells. A, B Circ_0007385 expression was examined using RT-qPCR in CRC tumor tissues as well as adjacent normal tissues. C Circ_0007385 expression in CRC cells and human normal colonic epithelial cell was detected by RT-qPCR. D RT-qPCR detected circ-MEMO1 and MEMO1 expression in CRC cells upon RNase R treatment. E RT-qPCR detected circ-MEMO1 expression in CRC cells after transfecting shRNAs targeting circ-MEMO1. F RT-qPCR detected MEMO1 expression in CRC cells after transfection of shRNAs targeting circ-MEMO1. G ENCORI database exhibited that MEMO1 expression was increased in COAD. H, I RT-qPCR detected MEMO1 expression in CRC tumor tissues along with adjacent normal tissues. J RT-qPCR detected MEMO1 expression in CRC cell lines and human normal colonic epithelial cell line. K FISH and IF assays verified the co-location of circ-MEMO1 and MEMO1 in CRC cells. **P < 0.01, ***P < 0.001
Silencing of Circ-MEMO1 Reduces the Malignant Behaviors in CRC Cells
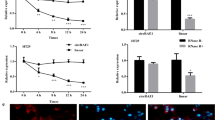
The biological regulation of circ-MEMO1 in CRC cells was explored. On the basis of colony formation assay, we found that circ-MEMO1 depletion inhibited CRC cells proliferation (Fig. 2A). Besides, TUNEL assay showed that of CRC cells apoptosis was elevated upon circ-MEMO1 silence (Fig. 2B, C). Moreover, transwell assays indicated that the migratory and invasive capacities of CRC cells were impaired when circ-MEMO1 was down-regulated (Fig. 2D). Epithelial-mesenchymal transition (EMT) is a modulatory program able to promote invasion as well as metastasis in cancer (García-Cuellar et al. 2021). Herein, through IF assay, it was also discovered that reduced expression of circ-MEMO1 could increase the fluorescence intensity of epithelial cell marker (E-cadherin) and decrease that of mesenchymal marker (N-cadherin), implying that circ-MEMO1 could accelerate the EMT process in CRC cells (Fig. 2E). All above data showed that circ-MEMO1 promoted CRC cell proliferation, migration, invasion, along with EMT.
Circ-MEMO1 silence reduces CRC cells malignant behaviors. A Colony formation assays assessed CRC cell proliferation after circ-MEMO1 silence. B, C TUNEL assays assessed CRC cell apoptosis after circ-MEMO1 silence. D Transwell assays assessed CRC cells migration and invasion after circ-MEMO1 silence. E IF assays assessed E-cadherin and N-cadherin expression in CRC cells after circ-MEMO1 silencing. ***P < 0.001
Oncogenic Function of Circ-MEMO1 in LoVo-Derived Xenograft Mouse Model of CRC
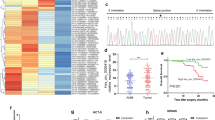
To investigate if knockdown of circ-MEMO1 affects CRC tumor growth and metastasis in vivo, NOD SCID immunodeficient mice received subcutaneously injection with circ-MEMO1-silenced and control LoVo cells. As revealed in Fig. 3A, after injection of LV-sh-circ-MEMO1, circ-MEMO1 expression in mice tumor tissues was definitely reduced. RT-qPCR further indicated that the transfection of LV-sh-circ-MEMO1 lessened circ-MEMO1 and MEMO1 expression, whereas elevated miR-485-3p/miR-543/miR-337-3p expression (Fig. 3B). Also, the tumor weight and tumor volume of mice were reduced after LV-sh-circ-MEMO1 transfection (Fig. 3C, D). Moreover, H&E staining results manifested that after circ-MEMO1 down-regulation, the tumor tissue developed larger areas of necrosis (Fig. 3E). In addition, TUNEL assay indicated that the cell apoptosis in mouse tumor was elevated after circ-MEMO1 depletion (Fig. 3F), and IF results showed that the EMT of CRC cells was weakened upon circ-MEMO1 reduction (Fig. 3G). All these data indicated that circ-MEMO1 accelerated CRC development and metastatic progression in vivo.
Oncogenic function of circ-MEMO1 in LoVo-derived xenograft mouse model of CRC. A RT-qPCR examined circ-MEMO1 expression in mouse tissues injected with LV-sh-circ-MEMO1-transfected LoVo cells. B RT-qPCR examined circ-MEMO1, MEMO1, and miR-485-3p/miR-543/miR-337-3p expression in xenograft tissues after circ-MEMO1 silencing. C, D Tumor weight and tumor tissues were assessed after circ-MEMO1 silencing. E H&E staining assessed the histopathological changes of tumor tissues after circ-MEMO1 silencing. F TUNEL assay assessed cell apoptosis in mice after circ-MEMO1 silence. G IF assessed E-cadherin together with N-cadherin expression in mouse tumor tissues after circ-MEMO1 silence. **P < 0.01, ***P < 0.001
Circ-MEMO1 Negatively Regulates MiR-485-3p/MiR-543/MiR-337-3p Expression
In view of that circ-MEMO1 and MEMO1 were mainly co-localized in the cytoplasm of CRC cells, it was conjectured that circ-MEMO1 might positively regulate MEMO1 expression through sponging miRNAs. Using ENCORI together with CircInteractome (https://circinteractome.nia.nih.gov/) databases, 3 miRNAs containing miR-485-3p/miR-543/miR-337-3p were found to bind with circ-MEMO1 and MEMO1 simultaneously (Fig. 4A). RT-qPCR results further manifested that overexpressed circ-MEMO1 could suppress the miR-485-3p/miR-543/miR-337-3p expression (Fig. 4B). Besides, dual-luciferase reporter assay demonstrated that circ-MEMO1 elevation apparently lessened the luciferase intensity of wild-type of miR-485-3p/miR-543/miR-337-3p, but had no impact on that of mutant-type of these miRNAs (Fig. 4C–E), which further confirmed the binding of circ-MEMO1 and these 3 miRNAs. Additionally, RIP assay also exhibited that circ-MEMO1 and these 3 miRNAs were more abundant after anti-Ago2 immunoprecipitation relative to IgG (Fig. 4F), which provided evidence that circ-MEMO1 and these 3 miRNAs occupied the same Ago2 protein to generate an RNA-induced silencing complex (RISC) in CRC cells. All these outcomes implied that circ-MEMO1 bound with miR-485-3p/miR-543/miR-337-3p.
Circ-MEMO1 negatively regulates miR-485-3p, miR-543, and miR-337-3p expression. A ENCORI and CircInteractome databases predicted that miR-485-3p/miR-543/miR-337-3p could bind with circ-MEMO1 and MEMO1. B MiR-485-3p/miR-543/miR-337-3p expression was examined by RT-qPCR in CRC cells after circ-MEMO1 silencing. C–E Dual-luciferase reporter assays detected the luciferase activity of miR-485-3p/miR-543/miR-337-3p WT and MUT in CRC cells after circ-MEMO1 silencing. F RIP assay detected the abundance of circ-MEMO1 and miR-485-3p/miR-543/miR-337-3p in Anti-Ago2. *P < 0.05, **P < 0.01, ***P < 0.001
Down-Regulation of MiR-485-3p/MiR-543/MiR-337-3p in CRC
MiR-485-3p/miR-543/miR-337-3p expression in CRC was explored, respectively. As revealed in Fig. 5A, miR-485-3p/miR-543/miR-337-3p expression was lower in CRC cells relative to human normal colonic epithelial cell line (HCoEpiC). Similarly, we discovered that these 3 miRNAs were underexpressed in CRC tumor tissues compared to adjacent normal tissues (Fig. 5B and E–F). Moreover, ENCORI database presented that miR-485-3p showed low expression in COAD (Fig. 5C). KM-plotter (https://kmplot.com/analysis/) database indicated that CRC patients with high miR-485-3p expression possessed longer survival time (Fig. 5D). All these data suggested that miR-485-3p/miR-543/miR-337-3p were down-regulated in CRC.
Down-regulation of miR-485-3p/miR-543/miR-337-3p in CRC. A RT-qPCR detected miR-485-3p/miR-543/miR-337-3p expression in CRC cells and human normal colonic epithelial cell. B and E, F RT-qPCR detected miR-485-3p/miR-543/miR-337-3p expression in CRC tumor tissues along with adjacent normal tissues. C ENCORI database predicted miR-485-3p expression in COAD. D KM-plotter analyzed relation of miR-485-3p expression and survival time of CRC patients. **P < 0.01, ***P < 0.001
MEMO1 is a Target of MiR-485-3p/MiR-543/MiR-337-3p
The relation of MEMO1 and miR-485-3p/miR-543/miR-337-3p was further probed. Firstly, miR-485-3p/miR-543/miR-337-3p expression was elevated in CRC cells by transfecting the indicated overexpression plasmid, respectively (Fig. 6A). Then, RT-qPCR together with western blot results exhibited that elevated miR-485-3p/miR-543/miR-337-3p expression lessened MEMO1 mRNA together with protein levels (Fig. 6B, C). Besides, RIP assay exhibited that MEMO1 together with miR-485-3p/miR-543/miR-337-3p were mainly enriched in miRNPs including Ago2 (Fig. 6D). Additionally, we found that miR-485-3p/miR-543/miR-337-3p overexpression could repress the luciferase intensity of MEMO1 3’UTR wild-type, but rarely affected that of MEMO1 3’UTR mutant-type (Fig. 6E). All these outcomes revealed that MEMO1 was targeted by miR-485-3p/miR-543/miR-337-3p.
MEMO1 is targeted by miR-485-3p/miR-543/miR-337-3p. A RT-qPCR detected miR-485-3p/miR-543/miR-337-3p expression in CRC cells after transfecting miR-485-3p mimic, miR-543 mimic, and miR-337-3p mimic. B, C RT-qPCR and western blot detected MEMO1 mRNA and protein expression in CRC cells after transfecting miR-485-3p mimic, miR-543 mimic, and miR-337-3p mimic. D RIP assay analyzed the enrichment of MEMO1 and miR-485-3p/miR-543/miR-337-3p in Anti-Ago2. E Dual-luciferase reporter assay detected the luciferase activity of MEMO1 3’UTR WT and MUT in CRC cells after overexpression of miR-485-3p/miR-543/miR-337-3p. *P < 0.05, **P < 0.01, ***P < 0.001
MiR-485-3p/MiR-543/MiR-337-3p Overexpression Reduces CRC Cells Malignant Behaviors
Subsequently, through gain-of function experiments, it was observed in Fig. 7A–D that, after transfection of miR-485-3p mimic, miR-543 mimic as well as miR-337-3p mimic in CRC cells, the cell proliferation, migration, invasion, as well as EMT process were hindered, while the cell apoptosis was elevated. All these data revealed that miR-485-3p/miR-543/miR-337-3p reduced CRC cell proliferation, migration, invasion, as well as EMT process.
MiR-485-3p/miR-543/miR-337-3p overexpression reduces CRC cell malignant behaviors. A Colony formation assays assessed CRC cell proliferation upon miR-485-3p/miR-543/miR-337-3p overexpression. B TUNEL assays assessed CRC cell apoptosis upon miR-485-3p/miR-543/miR-337-3p overexpression. C Transwell assays assessed CRC cell migration as well as invasion upon miR-485-3p/miR-543/miR-337-3p overexpression. D IF assays assessed E-cadherin and N-cadherin expression in CRC cells upon miR-485-3p/miR-543/miR-337-3p overexpression. ***P < 0.001
Circ-MEMO1 Promotes CRC Progression Via the MiR-485-3p/MiR-543/MiR-337-3p/MEMO1 Axis
To validate whether circ-MEMO1 regulated CRC cell malignancy in a miR-485-3p/miR-543/miR-337-3p- or MEMO1-dependent way, CRC cells were transfected with sh-NC, sh-circ-MEMO1, sh-circ-MEMO1 plus miR-485-3p/miR-543/miR-337-3p inhibitor and sh-circ-MEMO1 plus pcDNA-MEMO1, respectively. We found circ-MEMO1 silencing hindered cell proliferation, migration, invasion as well as EMT and elevated apoptotic cell numbers, whereas miR-485-3p/miR-543/miR-337-3p inhibition and MEMO1 overexpression recovered the effects caused by circ-MEMO1 silencing on CRC cells (Fig. 8A–E). These findings revealed that circ-MEMO1 functioned as an oncogene in CRC in a miR-485-3p/miR-543/miR-337-3p- or MEMO1-dependent manner.
Circ-MEMO1 facilitated CRC progression via the miR-485-3p/miR-543/miR-337-3p/MEMO1 axis. CRC cells were transfected with sh-NC, sh-circ-MEMO1, sh-circ-MEMO1 plus miR-485-3p/miR-543/miR-337-3p inhibitor and sh-circ-MEMO1 plus pcDNA-MEMO1, respectively. A Colony formation experiments tested cell proliferation. B TUNEL assays assessed cell apoptosis. C, D Transwell assays assessed cell migration as well as invasion. E IF assays assessed E-cadherin and N-cadherin expression. ***P < 0.001
Discussion
CRC is a prevalent gastrointestinal malignancy characterized by high morbidity and mortality (Sun et al. 2022). Accumulating evidence has validated that noncoding RNAs take part in CRC carcinogenesis (Zhang et al. 2022). CircRNAs have highly conserved and very stable circular structures, leading to them optimal biomarkers for diagnosis of CRC (Guo et al. 2021). For instance, Exosomal circPACRGL facilitates CRC progression via regulating TGF-β1 (Shang et al. 2020). CircMYH9 drives CRC growth by modulating serine metabolism as well as redox homeostasis in a p53-dependent way (Liu et al. 2021). CircRHOBTB3 impairs metastasis through affecting the HuR-mediated PTBP1 mRNA stability in CRC (Chen et al. 2021c). Here, we uncovered that circ-MEMO1, derived from the exon 3, 4, 5 loop of MEMO1 pre-mRNA, facilitated CRC progression via sponging miR-485-3p/miR-543/miR-337-3p to elevate MEMO1 expression (Graphical Abstract).
Our work demonstrated that circ-MEMO1 presented up-regulation in CRC. To the least of our knowledge, circ-MEMO1 expedites non-small cell lung cancer cell proliferation (Ding et al. 2022). Consistently, our experiments displayed that circ-MEMO1 depletion reduced the proliferation, migration, invasion along with EMT, while elevated the apoptosis of CRC cells. At same time, in vivo experiments also validated that circ-MEMO1 silencing markedly hindered the growth, metastasis, as well as EMT of CRC cells. All above outcomes strongly supported the oncogenic potential of circ-MEMO1 in CRC, which was consistent with former literatures (Ding et al. 2020).
To date, the modulation of circRNAs on their corresponding host genes belongs to a crucial method for circRNAs to exit their functions (Xu et al. 2020c). Jiayu Zhou et al. have pointed that circRNA-ENO1 increases tumor progression in lung adenocarcinoma through elevating ENO1 (Zhou et al. 2019). Xiaomin Li et al. have discovered that circITGA7 inhibits CRC metastasis via increasing ITGA7 transcription (Li et al. 2018). Hence, we further explored whether circ-MEMO1 affected CRC progression via regulating its host gene MEMO1. MEMO1 is related to EMT in breast cancer (Labrecque et al. 2021). Intriguingly, MEMO1 expression has been reported to be elevated in colon cancer cells (Xu et al. 2020b). Consistent with the above reports, our research also proved that MEMO1 was highly expressed in CRC. Notably, circ-MEMO1 could positively affect the expression of its host gene MEMO1 in CRC.
CircRNAs are generally thought to have miRNA-binding sites, which are in charge of miRNA relation and restore the restrictive role of miRNAs (Zhang et al. 2021b). As reported previously, circ-MEMO1 expedites non-small cell lung cancer cells proliferation via binding to miR-337-3p (Wei et al. 2022). Similarly, our study used bioinformatics analysis to further screen that miR-485-3p/miR-543/miR-337-3p bound to circ-MEMO1 and MEMO1, simultaneously. Further RIP and dual-luciferase reporter assays confirmed that circ-MEMO1 positively elevated MEMO1 expression via sponging miR-485-3p/miR-543/miR-337-3p. Substantial literature indicates that miRNAs are implicated in the regulation of diverse cellular processes and can serve as oncogenes or tumor suppressors in different malignancies (Kanwal et al. 2023). Former reports have also manifested that miR-485-3p and miR-543 work to be tumor repressor in the pathogenesis of CRC (Taherdangkoo et al. 2020). Besides, miR-337-3p has been documented to suppress breast cancer cells migration and invasion (Pan et al. 2021). Likewise, our study also indicated that miR-485-3p/miR-543/miR-337-3p was lowly expressed in CRC tissues and cells. Gain-of function assays further confirmed the suppressive roles of miR-485-3p/miR-543/miR-337-3p in CRC cell proliferation, migration, invasion, and EMT, which was in line with former reports (Jiang et al. 2021; Wang et al. 2020; Li and Li 2021). Additionally, rescue assays further demonstrated that circ-MEMO1 functioned as an oncogene in CRC in a miR-485-3p/miR-543/miR-337-3p- or MEMO1-dependent manner.
Conclusion
In summary, our study unveiled that a circRNA derived from the MEMO1 gene (circ-MEMO1) promoted CRC progression by regulating miR-485-3p/miR-543/miR-337-3p/MEMO1 axis. Thus, circ-MEMO1 was likely to be a promising therapeutic target for CRC.
Change history
09 August 2024
A Correction to this paper has been published: https://doi.org/10.1007/s10528-024-10900-8
References
Alavi M et al (2022) The efficiency of metal, metal oxide, and metalloid nanoparticles against cancer cells and bacterial pathogens: different mechanisms of action. Cell Mol Biomed Rep 2:10–21
Alsaedy H, Mirzaei AA, Alhashimi RAH (2022) Investigating the structure and function of long non-coding RNA (LncRNA) and its role in cancer. Cell Mol Biomed Rep. https://doi.org/10.2139/ssrn.4470175
Azizi Dargahlou S et al (2023) MicroRNAs; their therapeutic and biomarker properties. Cell Mol Biomed Rep 3(2):73–88
Bilal I et al (2021) Cytotoxic effect of diferuloylmethane, a derivative of turmeric on different human glioblastoma cell lines. Cell Mol Biomed Rep 1:14–22
Biller LH, Schrag D (2021) Diagnosis and treatment of metastatic colorectal cancer: a review. JAMA 325(7):669–685
Bin H et al (2022) The correlation between miR -34a-3p, miR -31, PLEK2 and the occurrence, development and prognosis of colorectal cancer. Cell Mol Biol (noisy-Le-Grand) 68(1):192–200
Chen RX et al (2019) Circular RNA circRNA_0000285 promotes cervical cancer development by regulating FUS. Eur Rev Med Pharmacol Sci 23(20):8771–8778
Chen K et al (2021a) Pathological features and prognostication in colorectal cancer. Curr Oncol 28(6):5356–5383
Chen L et al (2021b) The bioinformatics toolbox for circRNA discovery and analysis. Brief Bioinform 22(2):1706–1728
Chen J et al (2021c) Circular RNA circRHOBTB3 represses metastasis by regulating the HuR-mediated mRNA stability of PTBP1 in colorectal cancer. Theranostics 11(15):7507–7526
Chen C et al (2022) MicroRNA214 expression inhibits HCC cell proliferation through PTK2b/ Pyk2. Cell Mol Biol (noisy-Le-Grand) 68(1):20–25
Dekker E et al (2019) Colorectal cancer. Lancet 394(10207):1467–1480
Ding C et al (2020) Exosomal circ-MEMO1 promotes the progression and aerobic glycolysis of non-small cell lung cancer through targeting MiR-101-3p/KRAS axis. Front Genet 11:962
Ding D et al (2022) Circ_0007385 regulates cell proliferation, apoptosis and stemness via targeting miR-493-3p/RAB22A axis in non-small cell lung cancer. Thorac Cancer 13(4):571–581
García-Cuellar CM et al (2021) Particulate matter (PM(10)) promotes cell invasion through epithelial-mesenchymal transition (EMT) by TGF-β activation in A549 lung cells. Int J Mol Sci 22(23):12632
Guo Y et al (2021) Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR-30c-5p/TCF7 axis. Mol Cancer 20(1):93
He L, Zhou Y (2022) Evaluation of increased microRNA-21 in the serum of patients with cardia cancer. Cell Mol Biol (noisy-Le-Grand) 68(4):60–65
Jiang Z et al (2021) circ-Keratin 6c promotes malignant progression and immune evasion of colorectal cancer through microRNA-485-3p/programmed cell death receptor ligand 1 axis. J Pharmacol Exp Ther 377(3):358–367
Kanwal N et al (2023) Comprehensive analysis of microRNA (miRNA) in cancer cells. Cell Mol Biomed Rep 3(2):89–97
Kristensen LS et al (2022) The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol 19(3):188–206
Labrecque CL et al (2021) Identification of phenazine-based MEMO1 small-molecule inhibitors: virtual screening, fluorescence polarization validation, and inhibition of breast cancer migration. ChemMedChem 16(7):1163–1171
Li X et al (2018) Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J Pathol 246(2):166–179
Li C et al (2020) Integrated omics of metastatic colorectal cancer. Cancer Cell 38(5):734-747.e9
Li F et al (2021) Circular RNAs in cancer: limitations in functional studies and diagnostic potential. Semin Cancer Biol 75:49–61
Li L, Li Q (2021) miR-543 impairs breast cancer cell phenotypes by targeting and suppressing ubiquitin-conjugating enzyme E2T (UBE2T). Bioengineered 12(2):12394–12406
Li X, Mohammadi MR (2023) Combined diagnostic efficacy of red blood cell distribution width (RDW), prealbumin (PA), platelet-to-lymphocyte ratio (PLR), and carcinoembryonic antigen (CEA) as biomarkers in the diagnosis of colorectal cancer. Cell Mol Biomed Rep 3(2):98–106
Liu Z et al (2019) Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis 10(2):55
Liu X et al (2021) CircMYH9 drives colorectal cancer growth by regulating serine metabolism and redox homeostasis in a p53-dependent manner. Mol Cancer 20(1):114
Liu Z et al (2022) Circ_0022340 promotes colorectal cancer progression via HNRNPC/EBF1/SYT7 or miR-382–5p/ELK1 axis. Cell Mol Biol (noisy-Le-Grand) 68(7):107–116
Pan Y et al (2021) MiR-337-3p suppresses migration and invasion of breast cancer cells by downregulating ESRP1. Acta Histochem 123(7):151777
Saucedo-Sariñana AM et al (2022) Circulating cell-free-DNA concentration is a good biomarker for diagnosis of colorectal cancer in Mexican patients. Cell Mol Biol (noisy-Le-Grand) 68(6):1–8
Shang A et al (2020) Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol Cancer 19(1):117
Sun J et al (2022) Effects of mFOLFOX6 regimen combined with carrelizumab on immune function and prognosis in patients with microsatellite instability colorectal cancer. Cell Mol Biol (noisy-Le-Grand) 67(5):356–362
Tabin S et al (2022) Medical and medicinal importance of Rheum spp. collected from different altitudes of the Kashmir Himalayan range. Cell Mol Biomed Rep. https://doi.org/10.55705/cmbr.2022.349901.1050
Taherdangkoo K et al (2020) miR-485-3p suppresses colorectal cancer via targeting TPX2. Bratisl Lek Listy 121(4):302–307
Tourang M et al (2021) Association between Human Endogenous Retrovirus K gene expression and breast cancer. Cell Mol Biomed Rep 1:7–13
Wang Z et al (2020) miR-337-3p inhibits gastric tumor metastasis by targeting ARHGAP10. Mol Med Rep 21(2):705–719
Wei M et al (2022) Circ_0007385 promotes the proliferation and inhibits the apoptosis of non-small cell lung cancer cells via miR-337–3p-dependent regulation of LMO3. Histol Histopathol 38:797–810
Xu H et al (2020a) CircRNA_0000392 promotes colorectal cancer progression through the miR-193a-5p/PIK3R3/AKT axis. J Exp Clin Cancer Res 39(1):283
Xu K et al (2020b) miR-219a-1 inhibits colon cancer cells proliferation and invasion by targeting MEMO1. Cancer Biol Ther 21(12):1163–1170
Xu X et al (2020c) CircRNA inhibits DNA damage repair by interacting with host gene. Mol Cancer 19(1):128
Xu Y, Yang J (2022) The effect of 5A nursing combined with psychological nursing on the immune function, cancer-related fatigue and complications of patients undergoing radical resection of colorectal cancer. Cell Mol Biol (noisy-Le-Grand) 68(1):169–176
Yan H, Bu P (2021) Non-coding RNA in cancer. Essays Biochem 65(4):625–639
Yang J et al (2022) Hsa_circRNA_0088036 acts as a ceRNA to promote bladder cancer progression by sponging miR-140-3p. Cell Death Dis 13(4):322
Zhang W et al (2021a) CircRNA circFOXK2 facilitates oncogenesis in breast cancer via IGF2BP3/miR-370 axis. Aging (albany NY) 13(14):18978–18992
Zhang M et al (2021b) circRNA-miRNA-mRNA in breast cancer. Clin Chim Acta 523:120–130
Zhang F, Su T, Xiao M (2022) RUNX3-regulated circRNA METTL3 inhibits colorectal cancer proliferation and metastasis via miR-107/PER3 axis. Cell Death Dis 13(6):550
Zhou J et al (2019) CircRNA-ENO1 promoted glycolysis and tumor progression in lung adenocarcinoma through upregulating its host gene ENO1. Cell Death Dis 10(12):885
Zhou WY et al (2020) Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer 19(1):172
Zhu G et al (2021) CircRNA: a novel potential strategy to treat thyroid cancer (review). Int J Mol Med. https://doi.org/10.3892/ijmm.2021.5034
Funding
The authors have not disclosed any funding.
Author information
Authors and Affiliations
Contributions
JY: methodology, software; GL: resources, data curation; XZ: conceptualization, supervision, writing—reviewing and editing, Funding acquisition. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The author name Junfeng Yin has been corrected.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yin, J., Liu, G. & Zhu, X. MiR-485-3p/MiR-543/MiR-337-3p is Required for the Oncogenic Potential of the Hsa_circ_0007385-MEMO1 Axis in Colorectal Cancer. Biochem Genet 62, 1182–1199 (2024). https://doi.org/10.1007/s10528-023-10472-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-023-10472-z