Abstract
The balance of osteoblasts and marrow adipocytes from bone marrow mesenchymal stem cells (BM-MSCs) maintains bone health. Under aging or other pathological stimuli, BM-MSCs will preferentially differentiate into marrow adipocytes and reduce osteoblasts, leading to osteoporosis. Long non-coding RNA differentiation antagonizing non-protein coding RNA (DANCR) participates in the osteogenic differentiation of human BM-MSCs, but the mechanism by which DANCR regulates the osteogenic differentiation of human BM-MSCs has not been fully explained. We observed that DANCR and prospero homeobox 1 (PROX1) were downregulated during osteogenic differentiation of human BM-MSCs, while miR-1301-3p had an opposite trend. DANCR overexpression decreased the levels of alkaline phosphatase, RUNX2, osteocalcin, Osterix in BM-MSCs after osteogenic induction, but DANCR silencing had the opposite result. Moreover, DANCR sponged miR-1301-3p to regulate PROX1 expression. miR-1301-3p overexpression reversed the suppressive role of DANCR elevation on the osteogenic differentiation of human BM-MSCs. Also, PROX1 elevation abolished the promoting role of miR-1301-3p overexpression on the osteogenic differentiation of human BM-MSCs. In conclusion, DANCR suppressed the osteogenic differentiation of human BM-MSCs through the miR-1301-3p/PROX1 axis, offering a novel mechanism by which DANCR is responsible for the osteogenic differentiation of human BM-MSCs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteoporosis (OP) is a metabolic bone disease that increases bone fragility and increases the risk of fractures. It is characterized by the reduction of osteoblasts and the accumulation of marrow adipocytes in the bone marrow compartment [1]. Osteoblasts and marrow adipocytes are derived from the differentiation of bone marrow mesenchymal stem cells (BM-MSCs) [2]. Under normal conditions, the mutual balance between osteogenic and adipogenic differentiation of BM-MSCs is strictly controlled to maintain bone health [3]. However, BM-MSCs will preferentially differentiate into adipocytes under aging or other pathological stimuli, leading to increased bone marrow fat and progressive bone loss [4, 5]. However, the molecular mechanism of the disorder of differentiation of BM-MSCs into osteoblasts and adipocytes is still unclear.
Long non-coding RNAs (lncRNAs), a type of transcripts (>200 nucleotides), have been proved to participate in diverse biological processes [6]. The dysregulation of lncRNAs is linked to many human diseases, including OP [7]. For example, lncRNA X inactive specific transcript (XIST) facilitated OP development by suppressing the differentiation of BM-MSCs into osteoblasts [8]. lncRNA GAS5 mitigated OP through accelerating the differentiation of BM-MSCs into osteoblasts by elevating RUNX family transcription factor 2 (RUNX2) expression by adsorbing microRNA (miR)-498 [9]. lncRNA differentiation antagonizing non-protein coding RNA (DANCR) is located on human chromosome 4q12. DANCR had been uncovered as a potential target for some tumors [10]. Furthermore, the inhibition of DANCR decreased compression force-induced osteoclast formation of periodontal ligament cells [11]. Moreover, the silence of DANCR accelerated proliferation and differentiation of osteoblasts [12]. However, the mechanism by which DANCE regulates osteogenic differentiation has not been thoroughly interpreted.
Herein, we verified that DANCR exerted an inhibitory impact on the differentiation of human BM-MSCs into osteoblasts. In addition, DANCR sponged miR-1301-3p to elevate prospero homeobox 1 (PROX1) expression, thereby repressing the osteogenic differentiation of human BM-MSCs.
Materials and methods
Cell culture
Human BM-MSCs and 293T cells were bought from Procell (Wuhan, China). Human BM-MSCs were cultured in a complete medium for BM-MSCs (Catalog No: CM-H166, Procell). 293T cells were cultured in Dulbecco’s modified Eagle medium (DMEM) (Sigma, St Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS) (Sigma) and 1% penicillin/streptomycin (Sigma).
Induction of osteogenic differentiation
For osteogenic differentiation, human BM-MSCs were cultured in the complete medium for 48 h. Then, the complete medium was replaced with DMEM containing 10% FBS (Sigma), 100 U/mL penicillin (Sigma), 100 U/mL streptomycin (Sigma), 0.1 μM dexamethasone (Sigma), 10 mM β-glycerol phosphate, and 50 μM ascorbic acid (Sigma) and cultured for 21 days (d). The medium was changed every 3 d.
Cell transfection
Short hairpin RNA (shRNA) against DANCR (sh-DANCR), sh-PROX1, negative control (NC) shRNA (sh-NC), miR-1301-3p mimic and it matching NC (mimic NC), the precursor of miR-1301-3p (miR-1301-3p) and its complementary sequence (anti-miR-1301-3p), as well as their corresponding NC (miR-NC and anti-miR-NC) were synthesized by GenePharma (Shanghai, China). For pGLVU6-sh-DANCR (sh-DANCR) and pCMV-DANCR (oe-DANCR) generation, the sequence of sh-DANCR or DANCR was synthesized and cloned into the pGLVU6 (GenePharma, Shanghai, China) or pCMV (vector) plasmids (OriGene, Rockville, MD, USA). For pCMV-MIR-miR-1301-3p (miR-1301-3p) and pCMV-MIR-anti-miR-1301-3p (anti-miR-1301-3p) construction, the precursor of miR-1301-3p or its complementary sequence (anti-miR-1301-3p) was synthesized and cloned into the downstream of pCMV-MIR vector (OriGene). For pGLVU6-sh-PROX1 (sh-PROX1) or pcDNA-PROX1 construction, the sequence of sh-PROX1 or PROX1 was cloned into pGLVU6 (GenePharma) or pcDNA vectors (Addgene, Shanghai, China).
For the production stable cell lines, these lentivirus vectors were transfected into 293T cells using polyethylenimine (Ploysciencs, Warrington, PA, USA). Then, the lentivirus particles were used to infect with BM-MSCs and then selected with Neomycin (MedChem Express, Princeton, NJ, USA) or puromycin (MedChem Express). BM-MSCs cells were transiently transfected using Lipofectamine 3000 reagent (Thermo Fisher Scientific, Waltham, MA, USA).
Quantitative real-time polymerase chain reaction (RT-qPCR)
Total RNA extraction was conducted using Trizol Reagent (Thermo Fisher Scientific). RNA concentration was quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific) at A260/A280 nm. RNA integrity was assessed using agarose gel electrophoresis. Total RNA was reversely transcribed using a PrimeScript™ RT Master Mix (Takara, Dalian, China) or miRNA first-stand cDNA synthesis kit (GeneCopoeia, Guangzhou, China). qRT-PCR was executed using an SYBR Green (Takara). Relative expression was counted using the 2−ΔΔCt method, and β-actin or U6 small nuclear RNA (U6 snRNA) was used as an internal. All primer sequences were presented in Table 1.
Western blotting
Total protein isolation was executed using the RIPA lysis buffer (Beyotime, Shanghai, China). Total protein was separated by using 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). The membrane was then blocked in tris buffered saline tween containing 5% non-fat milk. Subsequently, the membranes were incubated with primary antibodies at 4 °C for overnight. After washing with tris buffered saline tween, the membranes were incubated with a secondary antibody. The blots were visualized with the enhanced chemiluminescence (Thermo Fisher Scientific). All antibodies were displayed as below: glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (ab8245, 1:1000), alkaline phosphatase (ALP) (ab83259, 1:500), RUNX family transcription factor 2 (RUNX2) (ab236639, 1:1000), osteocalcin (OCN) (ab133612, 1:1000), Osterix (ab94744, 1:500), PROX1 (ab124910, 1:1000), and goat anti-rabbit immunoglobulin G (IgG) (ab181603, 1:10000). These antibodies were purchased from Abcam (Cambridge, MA, USA). GAPDH was used as a loading control and goat anti-rabbit IgG was used as a secondary antibody.
Bioinformatics analysis
The targets of DANCR were predicted by LncBase V.2 (http://carolina.imis.athena-innovation.gr/diana_tools/web/), Jefferson (https://cm.jefferson.edu/rna22), and starbase3.0 (http://starbase.sysu.edu.cn/) databases. The base sequence of miR-1301-3p complementary to PROX1 was predicted with the starbase3.0 database.
Dual-luciferase reporter assay
The wild-type (wt) sequences of DANCR and PROX1 3′untranslated regions (UTR) and their mutant (mut) sequences were synthesized and inserted into the pMIR-REPORT vector (Applied Biosystems, Foster, CA, USA), respectively. 293T cells were co-transfected with mimic NC or miR-1301-3p mimic and a luciferase reporter with DANCR wt, DANCR mut, PROX1 3′UTR wt, or PROX1 3′UTR mut using Lipofectamine 3000 reagent (Thermo Fisher Scientific). The luciferase activities were detected using the luciferase reporter assay kit (Promega, Madison, WI, USA) in a TD20/20 Luminometer (Turner Biosystems, Sunnyvale, CA, USA).
RNA immunoprecipitation (RIP) assay
The specific binding DANCR or PROX1 and miR-1301-3p were verified by RIP assay with a Magna RIP kit (Millipore). After osteogenic induction, BM-MSCs were lysed with RIP lysis buffer. The lysate (100 μL) was incubated with RIP buffer containing magnetic beads conjugated with IgG antibody (ab109489, 1:100, Abcam) or argonaute-2 (Ago2) antibody (ab186733, 1:50, Abcam). Then, the RNA complex was extracted using the RNeasy Mini Kit (Qiagen, San Diego, CA, USA). RT-qPCR was conducted to assess the enrichment of DANCR or PROX1 and miR-1301-3p in RNA complexes.
Statistical analysis
The data come from 3 independent experiments and was displayed as mean ± standard deviation. The Statistics Package for Social Sciences (version 13.0, SPSS Inc., Chicago, IL, USA) was used for statistical analysis. The difference between 2 groups was assessed with unpaired Student’s t test. The differences among 3 or more groups were evaluated using one-way variance analysis (ANOVA) with Turkey’s post hoc test. P < 0.05 was deemed significantly different.
Results
DANCR was lowly expressed during the osteogenic differentiation of human BM-MSCs
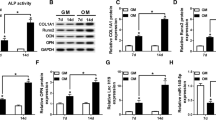
Previous research had uncovered that DANCR was related to the pathology of OP [13]. To verify the expression pattern of DANCR during osteogenic differentiation of human BM-MSCs, we performed RT-qPCR. As presented in Fig. 1a, DANCR expression was gradually decreased in human BM-MSCs at 7, 14, and 21 days after osteogenic induction. Meanwhile, the level of ALP protein (a phenotypic marker of osteogenic differentiation) was gradually elevated in BM-MSCs at 3, 7, 14, and 21 days after osteogenic induction (Fig. 1b). In addition, the levels of osteogenic differentiation-associated proteins (RUNX2, OCN, and Osterix) were increased in human BM-MSCs at 7, 14, and 21 days after osteogenic induction (Fig. 1c–e). These dada indicated that DANCR was downregulated during the osteogenic differentiation of human BM-MSCs.
DANCR was downregulated in human BM-MSCs during osteogenic differentiation. (a)–(e) Human BM-MSCs were cultured in osteogensis induction medium for 3, 7, 14, and 21 days. (a) RT-qPCR revealed the expression of DANCR in human BM-MSCs after osteogenic induction (n = 3, mean ± standard deviation). (b)–(e) Western blotting presented the levels of ALP, RUNX2, OCN, and Osterix in human BM-MSCs after osteogenic induction (n = 3, mean ± standard deviation). *P < 0.05
DANCR played a suppressive effect on the osteogenic differentiation of human BM-MSCs
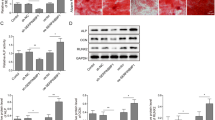
To explore the role of DANCR in the osteogenic differentiation of human BM-MSCs, we executed loss/gain-of-function experiments. The knockdown and overexpression efficiencies of sh-DANCR and oe-DANCR in human BM-MSCs were exhibited in Fig. 2a, b. Moreover, the silence of DANCR elevated the levels of ALP, RUNX2, OCN, and Osterix in human BM-MSCs after osteogenic induction (Fig. 2c). Reversely, the levels of ALP, RUNX2, OCN, and Osterix were decreased in DANCR-overexpressed human BM-MSCs after osteogenic induction (Fig. 2d). Together, these data indicated that DANCR repressed the osteogenic differentiation of human BM-MSCs.
Influence of DANCR on the osteogenic differentiation of human BM-MSCs. (a) and (b) RT-qPCR was conducted to validate the knockdown and overexpression efficiencies of sh-DANCR and oe-DANCR in human BM-MSCs (n = 3, mean ± standard deviation). (c) and (d) Human BM-MSCs were stably transfected with sh-DANCR, sh-NC, vector, or oe-DANCR and then cultured in osteogensis induction medium for 14 days. Impacts of DANCR silencing or overexpression on the levels of ALP, RUNX2, OCN, and Osterix in human BM-MSCs after osteogenic induction were assessed by western blotting (n = 3, mean ± standard deviation). *P < 0.05
DANCR was validated as a sponge for miR-1301-3p
To survey the regulatory mechanism of DANCR in the osteogenic differentiation of human BM-MSCs, we searched for miRs containing complementary sites to DANCR using Jefferson, starbase3.0, and LncBase V.2 databases. Overlapping data presented that there were 6 miRs (miR-374-3p, miR-1301-3p, miR-1914-3p, miR-6738-5p, miR-193b-3p, and miR-518d-3p) containing base sequences complementary to DANCR (Fig. 3a). Moreover, 4 miRs (miR-374-3p, miR-1301-3p, miR-6738-5p, and miR-193b-3p) could be regulated by DANCR in human BM-MSCs, especially miR-1301-3p (Fig. 3b, c). Also, miR-1301-3p was gradually upregulated at 3, 7, 14, and 21 days after osteogenic induction of human BM-MSCs (Fig. 3d). The binding sites between DANCR and miR-1301-3p were displayed in Fig. 3e. Furthermore, the luciferase activity of the DANCR wt reporter was reduced in 293T cells after co-transfection with miR-1301-3p mimic, but the luciferase activity of the DANCR mut reporter did not change (Fig. 3f). RIP assay exhibited that DANCR and miR-1301-3p were enriched in the anti-Ago2 group but not in the anti-IgG group, manifesting that DANCR and miR-1301-3p were co-existed in RNA-induced silencing complex (RISC) (Fig. 3g). These findings manifested that DANCR acted as a sponge for miR-1301-3p in human BM-MSCs.
DANCR adsorbed miR-1301-3p in human BM-MSCs. (a) The targets of DANCR were predicted by Jefferson, starbase3.0, and LncBase V.2 databases. (b) and (c) RT-qPCR presented the influence of DANCR inhibition or overexpression on the expression of miR-374-3p, miR-1301-3p, miR-1914-3p, miR-6738-5p, miR-193b-3p, and miR-518d-3p in human BM-MSCs (n = 3, mean ± standard deviation). (d) RT-qPCR revealed the expression of miR-1301-3p in human BM-MSCs after osteogenic induction for 3, 7, 14, and 21 days (n = 3, mean ± standard deviation). (e) The binding sites of DANCR on miR-1301-3p. (f) Dual-luciferase reporter assay was performed to verify the combination of DANCR and miR-1301-3p (n = 3, mean ± standard deviation). (g) RIP assay was performed using Ago2 and IgG antibodies (n = 3, mean ± standard deviation). *P < 0.05
DANCR regulated the osteogenic differentiation of human BM-MSCs via adsorbing miR-1301-3p
Based on the above findings, we further analyzed whether DANCR regulated the osteogenic differentiation of human BM-MSCs via sponging miR-1301-3p. The overexpression and knockdown efficiencies of miR-1301-3p and anti-miR-1301-3p were showed in Fig. 4a. Moreover, DANCR inhibition elevated miR-1301-3p expression in human BM-MSCs after osteogenic induction, while this tendency was restored after miR-1301-3p silencing (Fig. 4b). Inversely, miR-1301-3p overexpression abolished the downregulation of miR-1301-3p in DANCR-elevated human BM-MSCs after osteogenic induction (Fig. 4c). Also, the promoting effect of DANCR silencing on the levels of ALP, RUNX2, OCN, and Osterix in human BM-MSCs after osteogenic induction was offset by miR-1301-3p knockdown (Fig. 4d). In addition, the elevation of miR-1301-3p overturned the inhibitory influence of DANCR overexpression on the levels of ALP, RUNX2, OCN, and Osterix in human BM-MSCs after osteogenic induction (Fig. 4e). Collectively, these results suggested that DANCR modulated the osteogenic differentiation of human BM-MSCs via adsorbing miR-1301-3p.
DANCR adsorbed miR-1301-3p to modulate the osteogenic differentiation of human BM-MSCs. (a) Human BM-MSCs were stably transfected with miR-NC, miR-1301-3p, anti-miR-NC, or anti-miR-1301-3p. RT-qPCR exhibited the expression of miR-1301-3p in human BM-MSCs (n = 3, mean ± standard deviation). (b)–(e) Human BM-MSCs were stably transfected with sh-NC, sh-DANCR, sh-DANCR+anti-miR-NC, sh-DANCR+anti-miR-1301-3p, vector, oe-DANCR, oe-DANCR+miR-NC, or oe-DANCR+miR-1301-3p and then cultured in osteogensis induction medium for 14 days. (b) and (c) RT-qPCR was executed to analyze miR-1301-3p expression in human BM-MSCs after osteogenic induction (n = 3, mean ± standard deviation). (d) and (e) Western blotting was applied to detect the levels of ALP, RUNX2, OCN, and Osterix in human BM-MSCs after osteogenic induction (n = 3, mean ± standard deviation). *P < 0.05
PROX1 was verified as a target of miR-1301-3p
PROX1 had been identified as the main regulator of the lymphatic system phenotype [14]. To verify the biological function of PROX1 during osteogenic differentiation of human BM-MSCs, we detected the level of PROX1 protein during osteogenic differentiation of human BM-MSCs. The level of PROX1 protein was reduced in human BM-MSCs at 7, 14, and 21 days after osteogenic induction (Fig. 5a). Moreover, we discovered that PROX1 might be a target of miR-1301-3p through the starbase3.0 database (Fig. 5b). Also, miR-1301-3p mimic curbed the luciferase activity of the PROX1 3′UTR wt reporter in 293T cells, but there was no evident variation in the PROX1 3′UTR mut reporter (Fig. 5c). Furthermore, PROX1 and miR-1301-3p were co-existed in RISC induced by Ago2 antibody in human BM-MSCs (Fig. 5d). In addition, the inhibition of miR-1301-3p restored the downregulation of PROX1 protein in DANCR-silenced human BM-MSCs after osteogenic induction (Fig. 5e). On the contrary, the forcing expression of DANCR elevated PROX1 protein level in human BM-MSCs after osteogenic induction, but this increase was reversed by miR-1301-3p overexpression (Fig. 5f). These data indicated that DANCR adsorbed miR-1301-3p to regulate PROX1 expression in human BM-MSCs after osteogenic induction.
DANCR adsorbed miR-1301-3p to regulate PROX1 expression. (a) Western blotting revealed PROX1 protein level in human BM-MSCs after osteogenic induction for 3, 7, 14, and 21 days (n = 3, mean ± standard deviation). (b) The binding sites of PROX1 on miR-1301-3p were predicted by the starbase3.0 database. (c) The binding sites between PROX1 and miR-1301-3p was verified by dual-luciferase reporter assay (n = 3, mean ± standard deviation). (d) RIP assay was performed to survey whether PROX1 and miR-1301-3p could be co-precipitated in RISC (n = 3, mean ± standard deviation). (e) and (f) Human BM-MSCs were stably transfected with sh-NC, sh-DANCR, sh-DANCR+anti-miR-NC, sh-DANCR+anti-miR-1301-3p, vector, oe-DANCR, oe-DANCR+miR-NC, or oe-DANCR+miR-1301-3p and then cultured in osteogensis induction medium for 14 days. Western blotting was employed to survey the level of PROX1 protein in human BM-MSCs after osteogenic induction (n = 3, mean ± standard deviation). *P < 0.05
MiR-1301-3p regulated the osteogenic differentiation of human BM-MSCs via targeting PROX1
Given that miR-1301-3p targeted PROX1 in human BM-MSCs after osteogenic induction, we further explored whether miR-1301-3p regulated the osteogenic differentiation of human BM-MSCs through PROX1. We observed that PROX1 protein level was decreased in human BM-MSCs after sh-PROX1 transfection and was elevated in human BM-MSCs after pcDNA-PROX1 transfection (Fig. 6a). Moreover, PROX1 overexpression overturned the downregulation of PROX1 protein in miR-1301-3p-increased human BM-MSCs after osteogenic induction (Fig. 6b). Reversely, the silence of PROX1 abolished the upregulation of PROX1 protein in miR-1301-3p-inhibited human BM-MSCs after osteogenic induction (Fig. 6c). Furthermore, forced miR-1301-3p expression increased the levels of ALP, RUNX2, OCN, and Osterix in human BM-MSCs after osteogenic induction, but this elevation was abolished by the introduction of pcDNA-PROX1 (Fig. 6d). Also, the levels of ALP, RUNX2, OCN, and Osterix were reduced in miR-1301-3p-silenced human BM-MSCs after osteogenic induction, but this tendency was restored after PROX1 knockdown (Fig. 6e). These results indicated that miR-1301-3p modulated the osteogenic differentiation of human BM-MSCs via targeting PROX1.
MiR-1301-3p targeted PROX1 to modulate the osteogenic differentiation of human BM-MSCs. (a) The silencing and overexpression efficiencies of sh-PROX1 and pcDNA-PROX1 were verified by western blotting (n = 3, mean ± standard deviation). (b)–(e) Human BM-MSCs were stably transfected with miR-NC, miR-1301-3p, miR-1301-3p+pcDNA, miR-1301-3p+pcDNA-PROX1, anti-miR-NC, anti-miR-1301-3p, anti-miR-1301-3p+sh-NC, or anti-miR-1301-3p+sh-PROX1 and then cultured in osteogensis induction medium for 14 days. (b) and (c) Western blotting revealed the level of PROX1 protein in human BM-MSCs after osteogenic induction (n = 3, mean ± standard deviation). (d) and (e) Western blotting presented the levels of ALP, RUNX2, OCN, and Osterix in human BM-MSCs after osteogenic induction (n = 3, mean ± standard deviation). *P < 0.05
Discussion
Mounting evidence has confirmed that lncRNAs serve as important epigenetic regulatory factors of bone development and homeostasis [15]. Moreover, lncRNAs have been uncovered to take part in the differentiation of human BM-MSCs into osteoblasts [16]. Herein, we uncovered that DANCR exerted a repressive effect on the differentiation of human BM-MSCs into osteoblasts by elevating PROX1 expression via sponging miR-1301-3p, which offered a novel mechanism for understanding the osteogenic differentiation of human BM-MSCs.
Previous studies had proved that DANCR participated in osteogenic differentiation. Report of Wang et al. discovered that DANCR inhibition accelerated the differentiation of periodontal ligament stem cells into osteoblasts [17]. Moreover, DANCR activated the Wnt/β-catenin pathway via sponging miR-320a, thereby constraining the differentiation of BM-MSCs into osteoblasts [18]. Additionally, DANCR knockdown facilitated the differentiation of human BM-MSCs into osteoblasts via blocking the p38 MAPK pathway [19]. Herein, we verified that DANCR was lowly expressed during osteogenic differentiation of human BM-MSCs. After osteogenic induction, DANCR negatively modulated the levels of ALP, RUNX2, OCN, and Osterix in human BM-MSCs. These data indicated that DANCR constrained the differentiation of human BM-MSCs into osteoblasts.
According to the ceRNA hypothesis, lncRNAs can repress the activity of miRs through acting as miR molecular sponges [20]. MiRs can cause mRNA degradation or inhibit translation by recruiting RISC to the complementary sequence of target mRNAs [21]. Many studies have revealed that miRs are related to the differentiation of BM-MSCs into osteoblasts [22]. MiR-1301-3p had been proved to play an anti-tumor role in some cancers, such as papillary thyroid cancer [23], glioma [24], hepatocellular cancer [25], and esophageal squamous cell cancer [26]. STAT3-induced ABHD11-AS1 facilitated papillary thyroid cancer progression by upregulating STAT3 and activating the phosphoinosotide-3-kinase/v-akt murine thymoma viral oncogene homologue (PI3K/AKT) pathway via sponging miR-1301-3p [23]. Also, miR-1301-3p repressed glioma growth by repressing the Ras/MEK/ERK1/2 pathway by downregulating N-Ras [24]. In addition, miR-1301-3p curbed hepatocellular cancer progression via targeting BCL9 and blocking the Wnt/β-catenin pathway [25]. Also, miR-1301 could accelerate the differentiation of rat BM-MSCs into osteoblasts by targeting Satb2 expression [27]. Herein, DANCR was validated as a sponge for miR-1301-3p. Moreover, miR-1301-3p reversed DANCR-mediated influence on the protein levels of ALP, RUNX2, OCN, and Osterix in human BM-MSCs during osteogenic differentiation. Thus, we inferred that DANCR regulated the differentiation of human BM-MSCs into osteoblasts via adsorbing miR-1301-3p.
PROX1, a transcription factor, is related to cell fate determination and organ development [28, 29]. PROX1 has been uncovered as an oncogene or tumor suppressor according to different tumor types [30]. Furthermore, PROX1 overexpression induced the differentiation of human adipose-derived stem cells into lymphatic endothelial-like cells [31]. Igarashi et al. pointed out that vascular endothelial growth factor C (VEGF-C) decreased the expression of osteogenic differentiation marker genes by upregulating PROX1 and LYVE1 in transforming growth fector-beta (TGF-β)-responsive SG-2 cells (established mesenchymal stem cells) [32]. Herein, PROX1 was downregulated in human BM-MSCs during osteogenic differentiation. Moreover, PROX1 acted as a target for miR-1301-3p. Also, miR-1301-3p regulated the levels of ALP, RUNX2, OCN, and Osterix in human BM-MSCs through PROX1 during osteogenic differentiation. In addition, DANCR modulated PROX1 expression via sponging miR-1301-3p. Thus, we concluded that DANCR modulated the differentiation of human BM-MSCs into osteoblasts via regulating PROX1 expression via adsorbing miR-1301-3p. Unfortunately, whether DANCR directly affects the expression of PROX1 and the downstream pathways of the DANCR/miR-1301-3p/PROX1 axis have not been studied, which can be investigated in the future.
In conclusion, we verified that DANCR repressed the differentiation of human BM-MSCs into osteoblasts via elevating PROX1 expression through sponging miR-1301-3p. The research provided a novel mechanism for explaining the development of OP.
References
Letarouilly JG, Broux O, Clabaut A (2019) New insights into the epigenetics of osteoporosis. Genomics 111(4):793–798
Heino TJ, Hentunen TA (2008) Differentiation of osteoblasts and osteocytes from mesenchymal stem cells. Curr Stem Cell Res Ther 3(2):131–145
Kim J, Ko J (2014) A novel PPARγ2 modulator sLZIP controls the balance between adipogenesis and osteogenesis during mesenchymal stem cell differentiation. Cell Death Differ 21(10):1642–1655
Scheller EL, Rosen CJ (2014) What’s the matter with MAT? Marrow adipose tissue, metabolism, and skeletal health. Ann N Y Acad Sci 1311(1):14–30
Qadir A, Liang S, Wu Z, Chen Z, Hu L, Qian A (2020) Senile osteoporosis: the involvement of differentiation and senescence of bone marrow stromal cells. Int J Mol Sci 21(1):349
Peng WX, Koirala P, Mo YY (2017) LncRNA-mediated regulation of cell signaling in cancer. Oncogene 36(41):5661–5667
Wu QY, Li X, Miao ZN, Ye JX, Wang B, Zhang F, Xu RS, Jiang DL, Zhao MD, Yuan FL (2018) Long non-coding RNAs: a new regulatory code for osteoporosis. Front Endocrinol 9:587
Chen X, Yang L, Ge D, Wang W, Yin Z, Yan J, Cao X, Jiang C, Zheng S, Liang B (2019) Long non-coding RNA XIST promotes osteoporosis through inhibiting bone marrow mesenchymal stem cell differentiation. Exp Ther Med 17(1):803–811
Feng J, Wang JX, Li CH (2019) LncRNA GAS5 overexpression alleviates the development of osteoporosis through promoting osteogenic differentiation of MSCs via targeting microRNA-498 to regulate RUNX2. Eur Rev Med Pharmacol Sci 23(18):7757–7765
Jin SJ, Jin MZ, Xia BR, Jin WL (2019) Long non-coding RNA DANCR as an emerging therapeutic target in human cancers. Front Oncol 9:1225
Zhang X, Zhao Y, Zhao Z, Han X, Chen Y (2019) Knockdown of DANCR reduces osteoclastogenesis and root resorption induced by compression force via Jagged1. Cell Cycle 18(15):1759–1769
Jiang SY, Miao YX, Hirokazu T, Zhu SZ, Lu JS (2019) Effects of lncRNA DANCR on proliferation and differentiation of osteoblasts by regulating the Wnt/β-catenin pathway. Eur Rev Med Pharmacol Sci 23(13):5558–5566
Tong X, Gu PC, Xu SZ, Lin XJ (2015) Long non-coding RNA-DANCR in human circulating monocytes: a potential biomarker associated with postmenopausal osteoporosis. Biosci Biotechnol Biochem 79(5):732–737
Hong YK, Detmar M (2003) Prox1, master regulator of the lymphatic vasculature phenotype. Cell Tissue Res 314(1):85–92
Hassan MQ, Tye CE, Stein GS, Lian JB (2015) Non-coding RNAs: epigenetic regulators of bone development and homeostasis. Bone 81:746–756
Ju C, Liu R, Zhang YW, Zhang Y, Zhou R, Sun J, Lv XB, Zhang Z (2019) Mesenchymal stem cell-associated lncRNA in osteogenic differentiation. Biomed Pharmacother 115:108912
Wang Z, Huang Y, Tan L (2020) Downregulation of lncRNA DANCR promotes osteogenic differentiation of periodontal ligament stem cells. BMC Dev Biol 20(1):2
Wang CG, Hu YH, Su SL, Zhong D (2020) LncRNA DANCR and miR-320a suppressed osteogenic differentiation in osteoporosis by directly inhibiting the Wnt/β-catenin signaling pathway. Exp Mol Med 52(8):1310–1325
Zhang J, Tao Z, Wang Y (2018) Long non-coding RNA DANCR regulates the proliferation and osteogenic differentiation of human bone-derived marrow mesenchymal stem cells via the p38 MAPK pathway. Int J Mol Med 41(1):213–219
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146(3):353–358
Mohr AM, Mott JL (2015) Overview of microRNA biology. Semin Liver Dis 35(1):3–11
Wang J, Liu S, Li J, Zhao S, Yi Z (2019) Roles for miRNAs in osteogenic differentiation of bone marrow mesenchymal stem cells. Stem Cell Res Ther 10(1):197
Wen J, Wang H, Dong T, Gan P, Fang H, Wu S, Li J, Zhang Y, Du R, Zhu Q (2019) STAT3-induced upregulation of lncRNA ABHD11-AS1 promotes tumour progression in papillary thyroid carcinoma by regulating miR-1301-3p/STAT3 axis and PI3K/AKT signalling pathway. Cell Prolif 52(2):e12569
Zhi T, Jiang K, Zhang C, Xu X, Wu W, Nie E, Yu T, Zhou X, Bao Z, Jin X, Zhang J, Wang Y, Liu N (2017) MicroRNA-1301 inhibits proliferation of human glioma cells by directly targeting N-Ras. Am J Cancer Res 7(4):982–998
Yang C, Xu Y, Cheng F, Hu Y, Yang S, Rao J, Wang X (2017) miR-1301 inhibits hepatocellular carcinoma cell migration, invasion, and angiogenesis by decreasing Wnt/β-catenin signaling through targeting BCL9. Cell Death Dis 8(8):e2999
Zhang C, Xie L, Fu Y, Yang J, Cui Y (2020) lncRNA MIAT promotes esophageal squamous cell carcinoma progression by regulating miR-1301-3p/INCENP axis and interacting with SOX2. J Cell Physiol 235:7933–7944
Kong J, Wan LP, Liu ZM, Gao ST (2020) MiR-1301 promotes adipogenic and osteogenic differentiation of BMSCs by targeting Satb2. Eur Rev Med Pharmacol Sci 24(7):3501–3508
Petrova TV, Mäkinen T, Mäkelä TP, Saarela J, Virtanen I, Ferrell RE, Finegold DN, Kerjaschki D, Ylä-Herttuala S, Alitalo K (2002) Lymphatic endothelial reprogramming of vascular endothelial cells by the Prox-1 homeobox transcription factor. EMBO J 21(17):4593–4599
Risebro CA, Searles RG, Melville AAD, Ehler E, Jina N, Shah S, Pallas J, Hubank M, Dillard M, Harvey NL, Schwartz RJ, Chien KR, Oliver G, Riley PR (2009) Prox1 maintains muscle structure and growth in the developing heart. Development 136(3):495–505
Elsir T, Smits A, Lindström MS, Nistér M (2012) Transcription factor PROX1: its role in development and cancer. Cancer Metastasis Rev 31(3-4):793–805
Deng J, Dai T, Sun Y, Zhang Q, Jiang Z, Li S, Cao W (2017) Overexpression of Prox1 induces the differentiation of human adipose-derived stem cells into lymphatic endothelial-like cells in vitro. Cell Rep 19(1):54–63
Igarashi Y, Chosa N, Sawada S, Kondo H, Yaegashi T, Ishisaki A (2016) VEGF-C and TGF-β reciprocally regulate mesenchymal stem cell commitment to differentiation into lymphatic endothelial or osteoblastic phenotypes. Int J Mol Med 37(4):1005–1013
Funding
This study was supported by Zhejiang Province Public Welfare Technology Application Research Project (CN), China (Grant No. LGF20H060009).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Weng, W., Di, S., Xing, S. et al. Long non-coding RNA DANCR modulates osteogenic differentiation by regulating the miR-1301-3p/PROX1 axis. Mol Cell Biochem 476, 2503–2512 (2021). https://doi.org/10.1007/s11010-021-04074-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-021-04074-9