Abstract
Retinoblastoma (RB) is an intraocular malignancy that mainly occurs in infants and young children under 5 years of age. Circular RNA hsa_circ_0000034 (circ_0000034) was reported to be upregulated in RB tissues. Nevertheless, the function and mechanism of circ_0000034 in RB are unclear. Expression of circ_0000034, microRNA-361-3p (miR-361-3p), and a disintegrin and metalloproteinase 19 (ADAM19) was examined via quantitative real-time polymerase chain reaction (qRT-PCR). Cell viability, migration, invasion, and apoptosis were determined though Cell Counting Kit-8 (CCK-8), transwell, or flow cytometry assays. Caspase-3 activity was detected using a caspase-3 activity assay kit. Some protein levels were examined using Western blot analysis. Dual-luciferase reporter assay, RNA immunoprecipitation (RIP) assay, or RNA pull-down assay were performed to verify the relationship between circ_0000034 or ADAM19 and miR-361-3p. The function of circ_0000034 in vivo was confirmed via animal experiment. We verified that circ_0000034 expression was elevated in RB tissues and cells. Circ_0000034 silencing reduced RB growth in vivo, repressed viability, migration, invasion, and EMT, and induced apoptosis of RB cells in vitro. Circ_0000034 acted as a sponge for miR-361-3p, which targeted ADAM19 in RB cells. Furthermore, the inhibition of miR-361-3p restored circ_0000034 knockdown-mediated impacts on viability, migration, invasion, apoptosis, and EMT of RB cells. Moreover, ADAM19 overexpression abolished the influence of miR-361-3p mimic on viability, migration, invasion, apoptosis, and EMT of RB cells. Circ_0000034 expedited RB progression through upregulating ADAM19 via sponging miR-361-3p, which indicated that circ_0000034 might a target for RB therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Retinoblastoma (RB) is a common intraocular malignant tumor in children that originates from the retina embryonic nucleus cells [1]. Based on the preservation of children’s visual function, chemotherapy combined with adjuvant therapy has gradually become the preferred method for RB treatment [2]. However, this treatment method has side effects that can’t achieve the desired effect [3, 4]. Furthermore, some children have reached the advanced of RB at the time of diagnosis, which means that the possibility of eyeball removal is high [5]. Therefore, it is vital to explore the mechanism of RB advancement to develop the effective diagnostic markers and therapeutic targets for RB.
Circular RNAs (circRNAs), a new class of non-coding RNA, differ from linear RNA in that they have a covalently closed circular structure [6]. They are involved in regulating gene expression via modulating microRNAs (miRNAs), proteins, and transcription functions [7]. CircRNAs were revealed to be implicated in the tumorigenesis and advancement of tumors [8]. In recent, several studies revealed that circRNAs were associated with the malignant behaviors of RB [9,10,11]. For example, circRNA hsa_circ_0001649 regulated cell apoptosis and proliferation in RB through the AKT/mTOR pathway [9]. Circular RNA hsa_circ_0000034 (circ_0000034), located on chromosome 1:26,772,806–26,774,151, is derived from the DHDDS gene locus. RNA-sequencing analysis displayed that circ_0000034 was highly expressed in RB tissues [10]. However, the role of circ_0000034 in RB has not been reported.
MiRNAs, a class of single-stranded non-coding RNAs, bind to the 3′ Untranslated Region (UTR) of the target mRNAs to modulate gene expression [12]. They are involved in metabolism, immunity, development, differentiation, and cell death [13, 14]. MicroRNA-361-3p (miR-361-3p) was revealed to be associated with the advancement of some tumors, such as pancreatic ductal adenocarcinoma [15] and non-small cell lung cancer [16]. For instance, miR-361-3p modulated epithelial-mesenchymal transition (EMT) induced by ERK1/2 via DUSP2 in pancreatic ductal adenocarcinoma [15]. Moreover, miR-361-3p could regulate the stemness and proliferation of RB cells [17]. Nevertheless, the role and mechanism of miR-361-3p in RB have not been fully elucidated.
A disintegrin and metalloproteases (ADAMs) are a family of transmembrane proteins, which exert vital roles during cell migration, proteolysis, and adhesion [18]. The membrane-anchored ADAMs were responsible for the shedding of extracellular domains of cell surface proteins, such as receptors, cytokines, and potential forms of growth factors [19]. ADAM19 was uncovered to be associated with the proinflammatory and profibrotic processes in kidney disease [20]. ADAM19 was also revealed to be involved in the progression of tumors, such as glioma [21] and colorectal cancer [21]. Also, it was reported that ADAM19 was connected with the invasion and proliferation of RB cells [22]. Nevertheless, it is indistinct whether ADAM19 can be regulated by circ_0000034 and miR-361-3p in RB.
In consequence, we explored the role of circ_0000034 in RB. We also investigated the underlying mechanism of circ_0000034 in RB.
Materials and methods
Patients and specimens
This research was ratified by the Ethics Committee of the Fourth People’s Hospital of Shenyang. 30 paired RB tissues and neighboring normal tissues were collected from RB patients who underwent surgery at the Fourth People’s Hospital of Shenyang. Each participant’s informed consent was signed by their guardian prior to surgery. Exclusion criteria for this study was patients receiving other treatments before surgery.
Cell culture and transfection
Human retinal epithelial cells ARPE-19 and RB cell lines (Y79 and Weri-Rb1) were purchased from BeNa Culture Collection (Suzhou, China). RB cells SO-RB50 were obtained from Shanghai Qincheng Biological Technology Co. Ltd. (Shanghai, China). All cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Invitrogen, Carlsbad, CA, USA) complemented with fetal bovine serum (FBS, 10%, HyClone, Logan, UT, USA), streptomycin (100 μg/mL, Life Technologies, Grand Island, NY, USA), and penicillin (100 U/mL, Life Technologies) and grown in an incubator with 5% CO2 at 37 °C.
Short hairpin RNA (shRNA) targeting circ_0000034 (sh-circ#1, sh-circ#2, and sh-circ#3) and negative control (sh-NC) were procured from GenePharma (Shanghai, China). MiR-361-3p mimics and inhibitors (miR-361-3p and anti-miR-361-3p) and their negative controls (miR-NC and anti-NC) were obtained from GenePharma. The sequence of ADAM19 (NM_033274.5) was cloned into pcDNA3.0 vector (vector) (Invitrogen) to construct the overexpression vector for ADAM19 (ADAM19). Also, the oligonucleotides or vectors were transiently transfected into the cells by Lipofectamine 3000 reagent (Invitrogen) when RB cell confluence reached 80%.
Quantitative real-time polymerase chain reaction (qRT-PCR)
TRIzol reagent (Invitrogen) was employed to extract total RNA from RB tissues and cells. Total RNA (1 μg) was reversely transcripted into complementary DNA using a PrimeScript RT reagent Kit (Takara, Dalian, China) or miRNA First-Strand Synthesis Kit (Takara). QRT-PCR was executed using the SYBR Premix Ex Taq (Takara). Expression of circ_0000034, miR-361-3p, and ADAM19 was calculated through the 2−ΔΔCt method, and Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 small nuclear RNA (snRNA) was served as an internal control. The primers are presented in Table 1.
Cell counting Kit-8 (CCK-8) assay
Y79 and Weri-Rb1 cells were transfected with pointed oligonucleotides or vectors. Next, cells (5 × 104 cells/well) were seeded in 96-well plates and cultured for 48 h. Then, CCK-8 reagent (Beyotime, Shanghai, China) (10 μL) was added to each well and incubated for 2 h at 37 °C. A microplate reader (Bio-Tek, Winooski, VT, USA) was employed to analyze the optical density at 450 nm.
Transwell assay
The migration capacity of RB cells was evaluated using a transwell with 8 μm pore polycarbonate membrane (BD Biosciences, Bedford, MA, USA). In short, the pointed oligonucleotides or vectors were transfected into RB cells. Cells (5 × 104 cells) were seeded onto the top chamber containing RPMI 1640 medium (with 1% BSA for maintenance osmotic pressure). The lower chamber of the transwell was supplemented with RPMI 1640 medium (with 10% FBS as a chemoattractant). After culture for 24 h, the cells on the lower surface of the membrane were fixed with methanol (100%) and stained with crystal violet (0.25%, Sigma, Louis, Missouri, USA). The same method was applied to determine the invasion capacity of RB cells, but the transwell chamber was precoated with matrigel (BD Biosciences). In the end, the migrated or invaded cells were counted through a light microscope (Olympus, Tokyo, Japan).
Flow cytometry assay
The apoptosis of RB cells was analyzed using a Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (BD Biosciences). Briefly, after incubation for 48 h, transfected RB cells were collected and washed with PBS. Next, cells (1 × 105) were resuspended in binding buffer. Subsequently, Annexin V-FITC (10 μL) and PI (5 μL) were used to stain the cells for 15 min in the dark. The apoptotic rate of RB cells was assessed via the FACScan flow cytometry (BD Biosciences).
Caspase-3 activity assay
The activity of caspase-3 in transfected RB cells was analyzed with the caspase-3 activity assay kit (Beyotime). Caspase-3 activity assay was performed as previously described [23]. The optical density at 450 nm was detected via the microplate reader (Bio-Tek).
Western blot analysis
Total protein was extracted with RIPA lysis buffer (Beyotime). Western blot analysis was performed according to previous descriptions [23]. The protein bands were visualized through the ImmunoStar LD (Wako Pure Chemical, Osaka, Japan). The primary antibodies were listed as follows: rabbit anti-E-cadherin (E-cad) (ab40772, 1:10,000, Abcam, Cambridge, MA, USA), rabbit anti-N-cadherin (N-cad) (ab18203, 1:1000, Abcam), rabbit anti-ADAM19 (ab10480, 1:500, Abcam), and rabbit-anti-GAPDH (ab128915, 1:5000, Abcam). The secondary antibody goat anti-rabbit (ab97051, 1:10,000, Abcam) immunoglobulin G (IgG) was used in this research. GAPDH was regarded as a loading control.
Dual-luciferase reporter assay
The Circinteractome database or starBase database was employed for the prediction of the binding sites between miR-361-3p and circ_0000034 or ADAM19. The fragment of circ_0000034 (5′-ATCAGCAATGCTGTGAGAGAGATGGCCTGGGGGGTGGAGCAAG-3′) or 3′ UTR of ADAM19 (GeneBank: NM_033274.5; genomic DNA position 3661 to 4080) was synthesized and inserted into the Notl and Xhol sites in psiCHECK-2 vectors (Promega) to construct the circ_0000034-WT or ADAM19 3′UTR-WT luciferase reporters. The mutant circ_0000034-mutant (WT) and ADAM19 3′UTR-MUT reporters were constructed as the same method. RB cells were co-transfected with luciferase reporter vectors and miR-NC or miR-361-3p. Eventually, the luciferase intensity in RB cells was analyzed using the dual-luciferase reporter assay kit (Promega).
RNA immunoprecipitation (RIP) assay
The relationship between circ_0000034 and miR-361-3p was verified using the Magna RIP kit (Millipore, Bedford, MA, USA). In short, RB cells were lysed via RIP lysis buffer. Next, cell lysate was incubated in the RIP buffer harboring magnetic beads conjugated with anti-ago2 or anti-IgG antibodies (Millipore). Subsequently, the magnetic beads were incubated with proteinase K (Sigma) for the digestion of the proteins. The total RNA was isolated using the TRIzol reagent (Invitrogen). QRT-PCR was conducted to evaluate the abundance of circ_0000034 and miR-361-3p.
RNA pull-down assay
The Biotinylated (Bio)-NC and Bio-miR-361-3p were acquired from Sigma. Then, Bio-NC or Bio-miR-361-3p was transfected into RB cells, respectively. The Bio-coupled RNA complexes in the lysates of RB cells were pulled down through incubating with Dynabeads M-280 Streptavidin (Invitrogen) at 4 °C for 3 h. The beads were washed with lysis buffer (Beyotime) and the complexes were purified with TRIzol (Invitrogen). The enrichment of circ_0000034 was assessed through qRT-PCR.
Animal experiment
The animal experiment was ratified by the Ethics Committee of the Fourth People’s Hospital of Shenyang. BALB/c nude mice (athymic, 17–18 g, 5-week-old) were purchased from Shanghai Experimental Animal Center (Shanghai, China). Y79 cells with sh-NC or lentivirus-mediated sh-circ#1 were subcutaneously injected into the dorsal side of the nude mice (every 4 mice in a group) to establish the tumor xenograft models. The mice were kept under specific pathogen-free conductions and were free to obtain water and food. From day 7, the tumor volume was measured once a week and calculated by the equation: Volume = (length × width2)/2. The mice were euthanized under anesthesia on day 35 to take tumor tissues for subsequent analysis.
Statistical analysis
The experiments in vitro were executed in triplicate. Statistical analysis was conducted via GraphPad Prism 6.0 (GraphPad, San Diego, CA, USA) and SPSS 18.0 software (SPSS, Chicago, IL, USA). Data were exhibited as mean ± standard deviation. Statistical significance was evaluated by Student’s t test (the difference between 2 groups) or one-way variance analysis (ANOVA) (the differences among 3 or more groups). P < 0.05 was considered statistically significant.
Results
Circ_0000034 was upregulated in RB tissues and cells
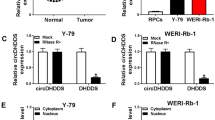
To verify the expression pattern of circ_0000034 in RB, we examined the expression level of circ_0000034 in 30 paired RB tissues and adjacent normal tissues. Circ_0000034 expression was elevated in RB tissues in comparison with neighboring normal tissues in the qRT-PCR analysis (Fig. 1a). Also, an overt elevation of circ_0000034 was observed in RB cells (Y79, Weri-Rb1, and SO-RB50) in contrast to that in the ARPE-19 cells (Fig. 1b). Moreover, circ_0000034 expression was higher in Y79 and Weri-Rb1 cells than that in SO-RB50 cells, so the Y79 and Weri-Rb1 cells were selected for subsequent studies. Therefore, these data indicated that elevated circ_0000034 expression might be implicated in the advancement of RB.
Circ_0000034 silencing repressed viability, migration, invasion, EMT, and accelerated apoptosis of RB cells
To survey the role of circ_0000034 in RB, we first designed three shRNA-targeting circ_0000034 (sh-circ#1, sh-circ#2, and sh-circ#3). Results of qRT-PCR displayed that circ_0000034 expression was overtly reduced in RB cells transfected with sh-circ#1, sh-circ#2, and sh-circ#3 compared to the sh-NC group (Fig. 2a). Furthermore, the sh-circ#1 was utilized to investigate the effects of circ_0000034 repression on viability, migration, invasion, apoptosis, and EMT of RB cells. CCK-8 assay presented that reduced circ_0000034 expression signally suppressed viability of Y79 and Weri-Rb1 cells (Fig. 2b). Transwell assay manifested that cell migration and invasion were observably curbed in Y79 and Weri-Rb1 cells after circ_0000034 inhibition (Fig. 2c, d). Flow cytometry assay exhibited that cell apoptotic proportion was distinctly increased in Y79 and Weri-Rb1 cells after transfection with sh-circ#1 (Fig. 2e, f). Moreover, the caspase-3 activity was higher in circ_0000034-silenced Y79 and Weri-Rb1 cells (Fig. 2g). Besides, the EMT-related proteins, E-cad and N-cad, were assessed by Western blot analysis. We discovered that circ_0000034 knockdown markedly elevated the level of E-cad and reduced the level of N-cad protein expression in both Y79 and Weri-Rb1 cells (Fig. 2h). Together, these results indicated that circ_0000034 acted as an oncogene in RB.
Impacts of circ_0000034 knockdown on viability, migration, invasion, apoptosis, and EMT of RB cells. a QRT-PCR was employed to evaluate circ_0000034 expression in Y79 and Weri-Rb1 cells transfected with sh-circ#1, sh-circ#2, sh-circ#3, or sh-NC. b–f Effect of circ_0000034 repression on viability, migration, invasion, and apoptosis of Y79 and Weri-Rb1 cells was detected through CCK-8, transwell, or flow cytometry assays. g The activity of caspase-3 in circ_0000034-silenced Y79 and Weri-Rb1 cells was detected through a caspase-3 activity assay kit. h The levels of E-cad and N-cad in circ_0000034-silenced Y79 and Weri-Rb1 cells were assessed via Western blot analysis. *P < 0.05
Circ_0000034 acted as a sponge for miR-361-3p in RB cells
To survey the molecular mechanism of circ_0000034 in RB, we predicted the possible target for circ_0000034 through Circinteractome database. The results exhibited that there were 10 miRNAs (miR-1184, miR-1204, miR-1205, miR-361-3p, miR-450b-3p, miR-769-3p, miR-1270, miR-620, miR-638, and miR-873) that possessed complementary sites to circ_0000034. We discovered that circ_0000034 silencing elevated the level of miR-361-3p in Y79 and Weri-Rb1 cells (Supplementary Fig. 1A and 1B). Thus, the miR-361-3p was selected for further investigation. The binding sites between miR-361-3p and circ_0000034 were shown in Fig. 3a. Dual-luciferase reporter assay manifested that the luciferase intensity of the circ_0000034-WT reporter was visibly decreased in Y79 and Weri-Rb1 cells transfected with miR-361-3p, while the luciferase intensity of the circ_0000034-MUT reporters did not change (Fig. 3b). Besides, RIP assay revealed that circ_0000034 and miR-361-3p were evidently enriched in ago2-containing beads in comparison with the control group (Fig. 3c). RNA pull-down assay showed that circ_0000034 could be pulled down by Bio-miR-361-3p probe relative to the Bio-NC probe (Fig. 3d). Additionally, circ_0000034 downregulation strikingly elevated miR-361-3p expression in Y79 and Weri-Rb1 cells (Fig. 3e). We also observed that miR-361-3p was lowly expressed in RB tissues than that in neighboring normal tissues (Fig. 3f). Collectively, these data suggested that circ_0000034 acted as a sponge for miR-361-3p in RB cells.
Circ_0000034 was verified as a sponge for miR-361-3p in RB cells. a The binding sites of miR-361-3p in circ_0000034 were predicted via Circinteractome database. b The luciferase activities of the circ_0000034-WT or circ_0000034-MUT reporters in Y79 and Weri-Rb1 cells transfected with miR-361-3p or miR-NC were analyzed through dual-luciferase reporter assay. c The relationship between miR-361-3p and circ_0000034 was further verified by RIP assay. d The enrichment of circ_0000034 by Bio-miR-361-3p or Bio-NC probes was assessed via qRT-PCR. e Effect of circ_0000034 downregulation on miR-361-3p expression of Y79 and Weri-Rb1 cells was analyzed via qRT-PCR. f Expression of miR-361-3p in 30 paired RB tissues and neighboring normal tissues was verified using qRT-PCR. *P < 0.05
Inhibition of miR-361-3p restored circ_0000034 silencing-mediated effects on viability, migration, invasion, apoptosis, and EMT of RB cells
Given that circ_0000034 targeted miR-361-3p in RB cells, we further verified whether circ_0000034 played its role in RB cells via miR-361-3p. We observed that the expression of miR-361-3p was prominently reduced in Y79 and Weri-Rb1 cells transfected with anti-miR-361-3p than that in the anti-NC group (Fig. 4a). CCK-8 assay revealed that miR-361-3p downregulation reversed the repression of viability of Y79 and Weri-Rb1 cells caused by circ_0000034 knockdown (Fig. 4b). Moreover, silenced miR-361-3p expression overturned the repressive impact of circ_0000034 repression on migration and invasion of Y79 and Weri-Rb1 cells (Fig. 4c, d). Also, the enhancement of apoptosis of Y79 and Weri-Rb1 cells induced by circ_0000034 inhibition was abolished after miR-361-3p silencing (Fig. 4e). In addition, miR-361-3p inhibition recovered the elevation of caspase-3 activity in circ_0000034-silenced Y79 and Weri-Rb1 cells (Fig. 4f). Furthermore, miR-361-3p silencing restored the influence of circ_0000034 knockdown on the levels of E-cad and N-cad in Y79 and Weri-Rb1 cells (Fig. 4g). In sum, these results demonstrated that circ_0000034 played a carcinogenic role in RB via miR-361-3p.
Circ_0000034 exerted its role in RB cells via miR-361-3p. a The expression of miR-361-3p in Y79 and Weri-Rb1 cells transfected with anti-miR-361-3p or anti-NC was assessed via qRT-PCR. b–e Impact of miR-361-3p repression on circ_0000034 silencing-mediated viability, migration, invasion, and apoptosis of Y79 and Weri-Rb1 cells was analyzed via CCK-8, transwell, or flow cytometry assays. (F) Influence of miR-361-3p knockdown on caspase-3 activity in circ_0000034-silenced Y79 and Weri-Rb1 cells was detected by a caspase-3 activity assay kit. g Western blot analysis was executed to assess the influence of miR-361-3p knockdown on E-cad and N-cad protein expression of circ_0000034-silenced Y79 and Weri-Rb1 cells. *P < 0.05
ADAM19 acted as a target for miR-361-3p in RB cells
In view of the above findings, we further explored the downstream target for miR-361-3p. StarBase database presented that 10 mRNAs (E2F2, JAK1, NOTCH2, ETV3, FZD4, CCND2, ADAM19, AKT2, MAPK4, ADAM10) might be downstream targets of miR-361-3p. QRT-PCR exhibited that miR-361-3p mimic repressed the expression of ADAM19 mRNA in Y79 and Weri-Rb1 cells (Supplementary Fig. 2A and B). The binding sites between miR-361-3p and ADAM19 mRNA are displayed in Fig. 5a. Results of dual-luciferase reporter assay revealed that miR-361-3p enhancement remarkably repressed the luciferase activity of the ADAM19 3′UTR-WT reporter in Y79 and Weri-Rb1 cells, while there was no difference in the luciferase activity of the ADAM19 3′UTR-MUT reporter (Fig. 5b). Moreover, ADAM19 mRNA and protein levels were reduced in miR-361-3p-overexpressed Y79 and Weri-Rb1 cells (Fig. 5c, d). In addition, the expression of ADAM19 mRNA was increased in RB tissues (Fig. 5e). Also, miR-361-3p enhancement repressed viability, migration, and invasion of Y79 and Weri-Rb1 cells, while this influence was abrogated by ADAM19 overexpression (Fig. 5f–h). Furthermore, forced ADAM19 expression overturned the promotive influence of miR-361-3p elevation on apoptosis of Y79 and Weri-Rb1 cells (Fig. 5i). ADAM19 overexpression reversed miR-361-3p mimic-mediated effects on caspase-3 activity and the levels of E-cad and N-cad in Y79 and Weri-Rb1 cells (Fig. 5j–l). Taken together, these data manifested that miR-361-3p regulated the malignancy of RB cells via targeting ADAM19.
ADAM19 was a downstream target of miR-361-3p in RB cells. a The binding sites of ADAM19 in miR-361-3p were predicted through starBase database. b Dual-luciferase reporter assay was executed to assess the luciferase intensity of the ADAM19 3′UTR-WT or ADAM19 3′UTR-MUT in Y79 and Weri-Rb1 cells transfected with miR-361-3p or miR-NC. c and d Influence of miR-361-3p overexpression on the levels of ADAM19 mRNA and protein was analyzed via qRT-PCR or western blot analysis. e Relative expression of ADAM19 mRNA in 30 paired RB tissues and neighboring normal tissues was examined using qRT-PCR. f–i Impacts of ADAM19 elevation on miR-361-3p upregulation-mediated viability, migration, invasion, and apoptosis of Y79 and Weri-Rb1 cells were assessed via CCK-8, transwell, or flow cytometry assays. j Caspase-3 activity assay kit was employed to determine the influence of ADAM19 upregulation on caspase-3 activity in miR-361-3p-enhanced Y79 and Weri-Rb1 cells. k and l Western blot analysis was performed to determine the effects of ADAM19 overexpression on miR-361-3p mimic-mediated effects on the levels of E-cad and N-cad protein in Y79 and Weri-Rb1 cells. *P < 0.05
Circ_0000034 regulated ADAM19 expression via miR-361-3p in RB cells
Knowing that circ_0000034 served as a target of miR-361-3p, which targeted ADAM19 in RB cells, we further verified whether circ_0000034 regulated ADAM19 expression via miR-361-3p. The results displayed that circ_0000034 knockdown reduced the levels of ADAM19 mRNA and protein in Y79 and Weri-Rb1 cells, while this decrease was overturned after anti-miR-361-3p introduction (Fig. 6a–d). These data disclosed that circ_0000034-modulated ADAM19 expression via sponging miR-361-3p in RB cells.
Inhibition of circ_0000034 decreased tumor growth in vivo
Considering that circ_0000034 played a carcinogenic role in RB in vitro, we further confirmed the role of circ_0000034 in vivo via constructing xenograft mouse models. Circ_0000034 downregulation repressed tumor volume and weight compared to the sh-NC group (Fig. 7a, b). Also, the expression of circ_0000034 and ADAM19 protein was overtly reduced in mice tumor tissues of the sh-circ#1 group in contrast to the sh-NC group, while miR-361-3p expression had an opposite tendency (Fig. 7c–e). These data manifested that circ_0000034 exerted a carcinogenic role in RB.
Circ_0000034 knockdown inhibited tumor growth in vivo. a Tumor volume was measured once a week from day 7. b Tumor weight was assessed on day 35. c–e The expression of circ_0000034, miR-361-3p, and ADAM19 protein was detected with qRT-PCR or Western blot analysis. f Schematic of the regulation mechanism of circ_0000034 in RB. *P < 0.05
Discussion
RB often occurs in infants and young children under 5 years of age. In developing countries, the mortality of RB accounted for about 50–70% of all cases [24]. As a new type of non-coding RNA, circRNAs have attracted attention because they are promising as prognostic and diagnostic biomarkers in some tumors [25]. Emerging researches have disclosed that circRNAs are key regulators of tumorigenesis via regulation of the malignant behaviors of cancer cells [26, 27]. For instance, circRNA circ_0001982 accelerated cell carcinogenesis by downregulating miR-143 in breast cancer [28]. Furthermore, circRNA LARP4 cured cell invasion and proliferation through elevating LATS1 expression via sponging miR-424-5p in gastric cancer [29]. In RB, circRNA circ_0001649 facilitated tumor cell apoptosis and repressed tumor cell proliferation through the AKT/mTOR pathway [9]. Jiao et al. revealed that reduced circRNA circ_0093996 expression promoted RB progression via the miR-183/PDCD4 axis [10]. In this study, we verified that circ_0000034 was upregulated in RB tissues, which was in line with the study of Jiao et al. [10]. Moreover, the inhibition of circ_0000034 reduced RB growth in vivo and induced apoptosis, suppressed viability, migration, invasion, and EMT of RB in vitro. EMT is the process by which epithelial cells transform into motile mesenchymal cells [30]. It is well known that EMT is essential for embryogenesis, wound healing, and malignant progression [31]. These results revealed that circ_0000034 exerted a carcinogenic role in RB.
Increased researches demonstrated that circRNAs regulated gene expression via serving as sponge for miRNAs [11, 28, 29]. MiR-361-3p had been proved as a suppressor in a range of cancers [15, 16]. Previous study uncovered that miR-361-3p overexpression cured metastasis and proliferation of non-small cell lung cancer via downregulating SH2B1 [32]. Another report revealed that circRNA circ-MYBL2 repressed the expression of miR-361-3p in cervical cancer, which accelerated cancer cell invasion, proliferation, and EMT [33]. Herein, we found that circ_0000034 served as a sponge for miR-361-3p. The silence of miR-361-3p abolished circ_0000034 repression-mediated impacts on viability, migration, invasion, apoptosis, and EMT of RB cells. These data demonstrated that circ_0000034 exerted its influence on RB progression via sponging miR-361-3p.
It was reported that ADAM19 was highly expressed in a variety of tumors [34,35,36,37]. Previous research exhibited that the invasiveness of human primary brain tumors was associated with high ADAM19 expression level and activity [35]. Moreover, miR-30c elevation could curb colon cancer growth by suppressing ADAM19 expression [34]. Also, miR-145 mimic repressed ADAM19 expression, which repressed EMT in glioblastoma [38]. Furthermore, miR-145 overexpression could inhibit the invasion and proliferation of RB cells via targeting ADAM19 [22]. Herein, we confirmed that ADAM19 was a target of miR-361-3p. ADAM19 was regulated by circ_0000034 via miR-361-3p in RB cells. In addition, ADAM19 elevation recovered miR-361-3p elevation-mediated influence on viability, migration, invasion, apoptosis, and EMT of RB cells. Hence, we concluded that circ_0000034 regulated cell viability, migration, invasion, apoptosis, and EMT in RB via miR-361-3p/ADAM19 axis (Fig. 7f). Unfortunately, using one cell line for in vivo experiments is also a limitation of this study, and other cell lines can be used to further confirm the role of circ_0000034 in the future.
In sum, circ_0000034 silencing repressed RB advancement through downregulating ADAM19 via sponging miR-361-3p, which provided a possible target for the treatment of RB.
Change history
21 January 2022
A Correction to this paper has been published: https://doi.org/10.1007/s11010-021-04311-1
References
de Jong MC, Kors WA, de Graaf P, Castelijns JA, Kivela T, Moll AC (2014) Trilateral retinoblastoma: a systematic review and meta-analysis. Lancet Oncol 15:1157–1167. https://doi.org/10.1016/s1470-2045(14)70336-5
Pritchard EM, Dyer MA, Guy RK (2016) Progress in small molecule therapeutics for the treatment of retinoblastoma. Mini Rev Med Chem 16:430–454. https://doi.org/10.2174/1389557515666150722100610
Errico A (2014) Cancer therapy: retinoblastoma–chemotherapy increases the risk of secondary cancer. Nat Rev Clin Oncol 11:623. https://doi.org/10.1038/nrclinonc.2014.155
Fabian ID, Onadim Z, Karaa E, Duncan C, Chowdhury T, Scheimberg I, Ohnuma SI, Reddy MA, Sagoo MS (2018) The management of retinoblastoma. Oncogene 37:1551–1560. https://doi.org/10.1038/s41388-017-0050-x
Gao J, Zeng J, Guo B, He W, Chen J, Lu F, Chen D (2016) Clinical presentation and treatment outcome of retinoblastoma in children of South Western China. Medicine (Baltimore) 95:e5204. https://doi.org/10.1097/md.0000000000005204
Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H (2015) Circular RNA: a new star of noncoding RNAs. Cancer Lett 365:141–148. https://doi.org/10.1016/j.canlet.2015.06.003
Ebbesen KK, Hansen TB, Kjems J (2017) Insights into circular RNA biology. RNA Biol 14:1035–1045. https://doi.org/10.1080/15476286.2016.1271524
Zhao ZJ, Shen J (2017) Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol 14:514–521. https://doi.org/10.1080/15476286.2015.1122162
Xing L, Zhang L, Feng Y, Cui Z, Ding L (2018) Downregulation of circular RNA hsa_circ_0001649 indicates poor prognosis for retinoblastoma and regulates cell proliferation and apoptosis via AKT/mTOR signaling pathway. Biomed Pharmacother 105:326–333. https://doi.org/10.1016/j.biopha.2018.05.141
Lyu J, Wang Y, Zheng Q, Hua P, Zhu X, Li J, Li J, Ji X, Zhao P (2019) Reduction of circular RNA expression associated with human retinoblastoma. Exp Eye Res 184:278–285
Du S, Wang S, Zhang F, Lv Y (2019) SKP2, positively regulated by circ_ODC1/miR-422a axis, promotes the proliferation of retinoblastoma. J Cell Biochem 121:322
Hammond SM (2015) An overview of microRNAs. Adv Drug Deliv Rev 87:3–14. https://doi.org/10.1016/j.addr.2015.05.001
Kabekkodu SP, Shukla V, Varghese VK, D'Souza J, Chakrabarty S, Satyamoorthy K (2018) Clustered miRNAs and their role in biological functions and diseases. Biol Rev 93:1955–1986. https://doi.org/10.1111/brv.12428
Ambros V (2004) The functions of animal microRNAs. Nature 431:350–355. https://doi.org/10.1038/nature02871
Hu J, Li L, Chen H, Zhang G, Liu H, Kong R, Chen H, Wang Y, Li Y, Tian F, Lv X, Li G, Sun B (2018) MiR-361-3p regulates ERK1/2-induced EMT via DUSP2 mRNA degradation in pancreatic ductal adenocarcinoma. Cell Death Dis 9:807. https://doi.org/10.1038/s41419-018-0839-8
Chen L, Nan A, Zhang N, Jia Y, Li X, Ling Y, Dai J, Zhang S, Yang Q, Yi Y, Jiang Y (2019) Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol Cancer 18:13. https://doi.org/10.1186/s12943-019-0943-0
Zhao D, Cui Z (2019) MicroRNA-361-3p regulates retinoblastoma cell proliferation and stemness by targeting hedgehog signaling. Exp Ther Med 17:1154–1162. https://doi.org/10.3892/etm.2018.7062
Reiss K, Saftig P (2009) The "a disintegrin and metalloprotease" (ADAM) family of sheddases: physiological and cellular functions. Semin Cell Dev Biol 20:126–137. https://doi.org/10.1016/j.semcdb.2008.11.002
Takeda S (2016) ADAM and ADAMTS family proteins and snake venom metalloproteinases: a structural overview. Toxins (Basel). https://doi.org/10.3390/toxins8050155
Melenhorst WB, van den Heuvel MC, Timmer A, Huitema S, Bulthuis M, Timens W, van Goor H (2006) ADAM19 expression in human nephrogenesis and renal disease: associations with clinical and structural deterioration. Kidney Int 70:1269–1278. https://doi.org/10.1038/sj.ki.5001753
Qi B, Newcomer RG, Sang Q-XA (2009) ADAM19/adamalysin 19 structure, function, and role as a putative target in tumors and inflammatory diseases. Curr Pharm Des 15:2336–2348
Sun Z, Zhang A, Jiang T, Du Z, Che C, Wang F (2015) MiR-145 suppressed human retinoblastoma cell proliferation and invasion by targeting ADAM19. Int J Clin Exp Pathol 8:14521–14527
Wang F, Li Y, Zhang Z, Wang J, Wang J (2019) SHCBP1 regulates apoptosis in lung cancer cells through phosphatase and tensin homolog. Oncol Lett 18:1888–1894
Jabbour P, Chalouhi N, Tjoumakaris S, Gonzalez LF, Dumont AS, Chitale R, Rosenwasser R, Bianciotto CG, Shields C (2012) Pearls and pitfalls of intraarterial chemotherapy for retinoblastoma. J Neurosurg Pediatr 10:175–181. https://doi.org/10.3171/2012.5.peds1277
Zhang Y, Liang W, Zhang P, Chen J, Qian H, Zhang X, Xu W (2017) Circular RNAs: emerging cancer biomarkers and targets. J Exp Clin Cancer Res 36:152. https://doi.org/10.1186/s13046-017-0624-z
Wang R, Zhang S, Chen X, Li N, Li J, Jia R, Pan Y, Liang H (2018) CircNT5E acts as a sponge of miR-422a to promote glioblastoma tumorigenesis. Cancer Res 78:4812–4825. https://doi.org/10.1158/0008-5472.can-18-0532
Li XN, Wang ZJ, Ye CX, Zhao BC, Li ZL, Yang Y (2018) RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res 37:325. https://doi.org/10.1186/s13046-018-1006-x
Tang YY, Zhao P, Zou TN, Duan JJ, Zhi R, Yang SY, Yang DC, Wang XL (2017) Circular RNA hsa_circ_0001982 promotes breast cancer cell carcinogenesis through decreasing miR-143. DNA Cell Biol 36:901–908. https://doi.org/10.1089/dna.2017.3862
Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y, Chen X, Zhu J (2017) Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer 16:151. https://doi.org/10.1186/s12943-017-0719-3
Lamouille S, Xu J, Derynck R (2014) Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 15:178–196. https://doi.org/10.1038/nrm3758
Dongre A, Weinberg RA (2019) New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol 20:69–84. https://doi.org/10.1038/s41580-018-0080-4
Chen W, Wang J, Liu S, Wang S, Cheng Y, Zhou W, Duan C, Zhang C (2016) MicroRNA-361-3p suppresses tumor cell proliferation and metastasis by directly targeting SH2B1 in NSCLC. J Exp Clin Cancer Res 35:76. https://doi.org/10.1186/s13046-016-0357-4
Wang J, Li H, Liang Z (2019) circ-MYBL2 serves as a sponge for miR-361-3p promoting cervical cancer cells proliferation and invasion. Onco Targets Ther 12:9957–9964. https://doi.org/10.2147/ott.s218976
Zhang Q, Yu L, Qin D, Huang R, Jiang X, Zou C, Tang Q, Chen Y, Wang G, Wang X, Gao X (2015) Role of microRNA-30c targeting ADAM19 in colorectal cancer. PLoS ONE 10:e0120698. https://doi.org/10.1371/journal.pone.0120698
Wildeboer D, Naus S, Amy Sang QX, Bartsch JW, Pagenstecher A (2006) Metalloproteinase disintegrins ADAM8 and ADAM19 are highly regulated in human primary brain tumors and their expression levels and activities are associated with invasiveness. J Neuropathol Exp Neurol 65:516–527. https://doi.org/10.1097/01.jnen.0000229240.51490.d3
Wang Y, Lian YM, Ge CY (2019) MiR-145 changes sensitivity of non-small cell lung cancer to gefitinib through targeting ADAM19. Eur Rev Med Pharmacol Sci 23:5831–5839. https://doi.org/10.26355/eurrev_201907_18323
Chan MW, Huang YW, Hartman-Frey C, Kuo CT, Deatherage D, Qin H, Cheng AS, Yan PS, Davuluri RV, Huang TH, Nephew KP, Lin HJ (2008) Aberrant transforming growth factor beta1 signaling and SMAD4 nuclear translocation confer epigenetic repression of ADAM19 in ovarian cancer. Neoplasia 10:908–919. https://doi.org/10.1593/neo.08540
Wang X, Wang E, Cao J, Xiong F, Yang Y, Liu H (2017) MiR-145 inhibits the epithelial-to-mesenchymal transition via targeting ADAM19 in human glioblastoma. Oncotarget 8:92545–92554. https://doi.org/10.18632/oncotarget.21442
Funding
None.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no financial conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary file1 (TIF 275 kb)
Supplementary Fig.1 Influence of circ_0000034 inhibition on the predicted 10 miRNA levels in RB cells. (A and B) QRT-PCR revealed the levels of miR-1184, miR-1204, miR-1205, miR-361-3p, miR-450b-3p, miR-769-3p, miR-1270, miR-620, miR-638, and miR-873 in Y79 and Weri-Rb1 cells transfected with sh-circ#1 or sh-NC. *P < 0.05.
Supplementary file2 (TIF 281 kb)
Supplementary Fig.2 Influence of miR-361-3p overexpression on the predicted 10 mRNA levels in RB cells. (A and B) QRT-PCR revealed the levels of E2F2, JAK1, NOTCH2, ETV3, FZD4, CCND2, ADAM19, AKT2, MAPK4, ADAM10 mRNA in Y79 and Weri-Rb1 cells transfected with miR-361-3p or miR-NC. *P < 0.05.
Rights and permissions
About this article
Cite this article
Jiang, Y., Xiao, F., Wang, L. et al. Circular RNA has_circ_0000034 accelerates retinoblastoma advancement through the miR-361-3p/ADAM19 axis. Mol Cell Biochem 476, 69–80 (2021). https://doi.org/10.1007/s11010-020-03886-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-020-03886-5