Abstract
Purpose
Retinoblastoma (Rb) is the most common intraocular tumor in children. MicroRNAs (miRNAs) play a crucial role in gene regulation and cell growth/apoptosis/differentiation. The current study aimed to investigate the role of miR-498 in Rb.
Methods
Quantitative real-time polymerase chain reaction (QRT-PCR) was used to test mRNA level of miR-498. http://www.targetscan.org and http://www.microrna.org were applied to predict target of miR-498. Dual-luciferase reporter assay was applied to investigate if miR-498 targeted cell cycle progression 1 (CCPG1). Western blot (WB) was carried out to assess CCPG1 protein levels. 3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2-H-tetrazolium bromide (MTT) assay was used to evaluate cell proliferation. Annexin-V Fluorescein (FITC) was adopted to explore cell apoptosis.
Results
In Y79 cells, miR-498 was higher than in normal ARPE-19 cells. MiR-498 could recognize CCPG1-3′ untranslated region (UTR). CCPG1 protein level was remarkably decreased when overexpressed miR-498, nevertheless, significantly increased when inhibiting miR-498. Y79 cells that were transfected with miR-498 mimics manifested notable cell apoptosis down-regulation and cell proliferation promotion; whereas, those transfected with miR-498 inhibitor displayed significant cell apoptosis up-regulation and cell proliferation inhibition compared with control group.
Conclusion
Taken together, miR-498 promotes cell proliferation and inhibits cell apoptosis in Rb by directly targeting CCPG1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Retinoblastoma (Rb) is a rare form of cancer and initiates from immature cells in the retina, which nearly exclusively found in young children and whose most common and obvious sign is leukocoria [1]. Rb occurs unilaterally in two thirds of children or bilaterally in the remaining one-third [22], whose prevalence is approximately 1:15,000–1:20,000, accounts for 2–4% of all childhood malignancies [6, 23]. Some children with Rb even develop squint [13]. Rb occurs sporadically (60%) or can be inherited (40%) autosomal dominantly [3, 7, 21]; both are correlated with somatic function loss of tumor suppressor gene RB1 alleles. Rb has been the prototypic “model” cancer since Knudson’s comparison on the diagnosis age of bilateral vs. unilateral patients, which generated the “two-hit” hypothesis for cancer initiation [19]. Despite apparently enough to induce a tumor, these events could be modulated by multiple genetic changes in Rb. Mutation of genes in chromosomes can affect the way how cells grow and develop [12]. Alterations in RB1 or MYCN give rise to Rb. Researches on retinal cells demonstrate that two mutational events were not enough to initiate malignant transformation [8].
More than 10% human genome was regulated in a cell cycle-dependent manner [18]. miRNAs, a class of endogenous, small non-coding, regulatory, single-stranded RNAs of 18–25 nucleotides in length, negatively modulate gene expression at a posttranscriptional level by binding to the 3′UTR of target mRNA, ultimately generating mRNA degradation and protein activity reduction [2, 4, 11]. Consequently, miRNAs play a crucial role in gene regulation and cell growth/apoptosis/differentiation. Aberrant expression of miRNAs has been reported to participate in tumor initiation, progression, and invasion [14, 29].
Hsa-miR-498 was identified by Bentwich I et al. in 2005 [5]. The role of miR-498 in tumor development was demonstrated to be tissue-dependent; down-regulation of miR-498 was found in ovarian cancer [17, 24], non-small cell lung cancer [26], and colon cancer patients who were at the stage II [9]. However, miRNA microarray experiments and northern blot experiments demonstrated that hsa-miR-498 was elevated in human Rb and retinal tissues [30].
The current study aimed to elucidate the role of miR-498 in Rb.
Materials and methods
Cell culture
Y79 cells (Rb cell line) were cultured in RPMI-1640 medium supplemented with 20% fetal bovine serum (FBS), 50 ng/mL streptomycin, and 1.25 ng/mL amphotericin B at the temperature of 37 °C with 95% humidity and 5% CO2.
ARPE-19 cells (human normal epithelial cells of the retina) were cultured in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% FBS, 1% streptomycin/penicillin at an incubator with the temperature of 37 °C, and 5% CO2.
QRT-PCR
MiRNA was isolated from Y79 cells by virtue of mirVana miRNA isolation kit (Ambion, AM1560). After reverse transcription, QRT-PCR was performed in 96-well plates with Prism 7500 system (Lab India Instruments, Gurgaon, India). Reaction was as followed: 0.5 μg for cDNA, forward primer, reverse primer, and SYBR green reagent. Relative amount of miR498 was counted by 2–ΔΔCt (cycle number when amplification reaches the threshold). Primers were as followed: miR-498, forward: 5′-TTTCAAGCCAGGGGGCGTTTTTC-3′; reverse: 5′-GCTTCAAGCTCTGGAGGTGCTTTTC-3′; U6, forward: 5′-AACGCTTCACGAATTTGCGT-3′, reverse: 5′-CTCGCTTCGGCAGCACA-3′.
Prediction of target genes
Potential targets of miR-498 were predicted with database algorithm of miRBase (http://microrna.sanger.ac.uk) and TargetScan (http://genes.mit.edu/targetscan).
MiRNA transfection
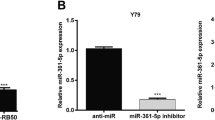
Y79 cells were seeded into 12-well plates at the concentration of 2 × 105 cells/well and incubated for about 24 h. Transfection of miR-498 mimics or inhibitor or negative control (NC) was carried out by Lipofectamine 2000, then incubated for about 48 h with OPTI-MEM I reduced serum medium (Invitrogen, CA, USA).
Cell proliferation analysis
At 48 h after transfection, Y79 cells were seeded into 96-well plates at the density of 5 × 103 cells/well. Into each well, 10 μl MTT solution was added. Y79 cells were incubated for another 4 h. Absorbance at 570 nm was read with microplate reader (Bio-Rad, CA, USA).
Cell apoptosis analysis
Cell apoptosis was assessed by fluorescein isothiocyanate (FITC) kit (Oncogene Research, San Diego, CA, USA) in accordance with the manufacturer’s instructions. Cell samples were analyzed by flow cytometry apparatus (Becton Dickinson FACSVantage SE, San Jose, CA, USA). Finally, cell number in each category was presented.
Dual-luciferase reporter assay
Y79 cells were seeded into 48-well plates. Twenty-four hours later, transfection was done with Lipofectamine 2000 and Opti-MEM I reduced serum medium (Invitrogen) with the total volume of 0.6 mL. Luciferase content was assessed at the initial 10 s of the reaction; firefly and Renilla luciferase activity were measured by dual-luciferase reporter assay system (Promega, Madison, WI) according to the manufacturer’s instructions and a luminometer (Molecular Device, Sunnyvale, CA).
WB
Cells were lysed in 0.5 mL radio-immunoprecipitation (RIPA) buffer. Lysates were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), blotted onto poly-vinylidene fluoride (PVDF) membranes. Afterwards, membranes were treated with primary antibody of CCPG1 or β-actin at 4 °C overnight. After rinse with Tris-buffered saline and Tween 20 (TBST) for three times, membranes were treated with horseradish peroxidase (HRP)-conjugated secondary antibody at room temperature for about 1 h. Bands were visualized by chemi-luminescence (ECL) kit. And β-actin (Sigma Aldrich, USA) acted as an endogenous control.
Statistical analysis
Data were presented as means ± standard deviation (SD) and analyzed by Student’s t test. Analyses were conducted by Statistic Package for Social Science (SPSS) 12.0 software. P < 0.05 presents significant difference.
Results
mRNA level of miR-498
mRNA level of miR-498 in Y79 cells and ARPE-19 cells were tested by RT-QPCR. In Y79 cell line, miR-498 was significantly higher when compared with normal cell. (Fig. 1) Taken together, the findings suggested that miR-498 level was associated with Rb.
Predicted target of miR-498
http://www.targetscan.org and http://www.microrna.org were applied to predict target miRNAs and Rb-related target genes, genes which showed higher grade in two softwares were selected with priority. We predicted that miR-498 could recognize CCPG1-3′ UTR (Fig. 2).
Interaction between miR-498 and CCPG1
Dual-luciferase reporter assay showed that miR-498 significantly inhibited the fluorescence of CCPG1-wild-type (WT) group compared with negative control (NC) group (Fig. 3a); nonetheless, there was no notable difference in fluorescence between CCPG1-mutant (MUT) group and NC group as displayed in Fig. 3b.
CCPG1 protein levels in different groups
Y79 cells were transfected with miR-498 mimics or inhibitor in experimental groups. CCPG1 protein levels in different groups were tested via western blot. CCPG1 was decreased after miR-498 overexpression and significantly increased when inhibiting expression of miR-498 (Fig. 4a). The corresponding statistical data were exhibited (Fig. 4b). These results indicate that miR-498 exerts an inhibitory effect on CCPG1 expression via interaction with its 3′-UTR in Y79 cells.
Y79 cell apoptosis followed by up-/down-regulation of miR-498
Y79 cells that were transfected with miR-498 NC showed no obvious cell apoptosis (Fig. 5a), miR-498 mimics led to a less cell apoptosis down-regulation (Fig. 5b), and miR-498 inhibitor exhibited significant cell apoptosis up-regulation compared with NC group (Fig. 5c). The statistical data were displayed in Fig. 5d.
Y79 cell apoptosis after miR-498 mimics/inhibitor transfection. a There was almost no cell apoptosis in Y79 cells that were transfected with miR-498 NC. b There was lower cell apoptosis in Y79 cells that were transfected with miR-498 mimics. c There was notable higher cell apoptosis in Y79 cells that were transfected with miR-498 inhibitor than in NC. d The corresponding statistical data were analyzed. **P < 0.01. N.S indicates no significant difference
Y79 cell proliferation followed by up-/down-regulation of miR-498
Y79 cells that were transfected with miR-498 mimics exhibited notably promoted cell proliferation (Fig. 6a); Y79 cells that transfected with miR-498 inhibitor displayed significantly inhibited cell proliferation compared with control group (Fig. 6b).
Discussion
Transcription factors (TFs) and miRNAs are crucial regulators for gene expression [20]. TFs promote or suppress gene transcription via binding with upstream regions of genes in a single form or collaborate with other proteins. While miRNAs participated in a large quantity of physiological and pathological processes including cancer [25, 28], miRNAs acted as either an oncogene or a tumor suppressor [16, 27]. miRNAs are commonly deregulated in cancers [10] via multiple mechanisms, for instance, dysfunction of key proteins in miRNA biogenesis pathway, mutation, and transcriptional deregulation and abnormality of DNA copy number. Literatures for miR-498 in cancers are relatively limited.
For instance, lower miR-498 expression is correlated with poorer prognosis in ovarian cancer [17], miR-498 inhibited the proliferation of human ovarian cancer cells [24], and miR-498 is down-regulated in non-small cell lung cancer [26] and colon cancer patients who were at the stage II [9]; however, it is up-regulated in human Rb and retinal tissues [30]. It is important to elucidate the role of hsa-miR-498 in Rb and to further identify miRNA-driven pathways.
We first carried out QRT-PCR to test whether there are differences in mRNA level of hsa-miR-498 between Y79 cells and ARPE-19 cells. Results showed that, in Y79 cell line, miR-498 was significantly higher than that in ARPE-19 cells, which was in line with the previous study [30].
Thereafter, we predicted the target gene of miR-498 and interaction site between miR-498 and its target gene. http://www.targetscan.org and http://www.microrna.org showed that miR-498 could recognize CCPG1-3′ UTR. Dual-luciferase reporter assay demonstrated that miR-498 indeed interacted with CCPG1-3′ UTR.
As the target gene of miR-498, CCPG1 was reported to play multiple biological functions. Knockdown of CCPG1 was reported to decrease cell viability, and natural antisense transcript of CCPG1 functionally destroys signaling pathways and corresponding biological phenotypes, including cell viability independently or in parallel with its sense transcript [15]. We further investigated whether CCPG1 expression level was affected by miR-498 activation/inhibition in Y79 cells.
WB showed that CCPG1 was remarkably decreased when overexpressed miR-498 significantly increased when inhibiting miR-498 expression. The data indicated that CCPG1 was targeted by miR-498. We are eager to know the effects of miR-498 activation/inhibition on cell apoptosis and proliferation in Y79 cells.
Y79 cells that were transfected with miR-498 mimics manifested notable down-regulated cell apoptosis and promoted cell proliferation; whereas, those transfected with miR-498 inhibitor displayed significant up-regulated cell apoptosis and inhibited cell proliferation compared with control group.
Collectively, we showed that miR-498 was up-regulated and its target CCPG1 was down-regulated in Y79 cell line. miR-498 increases cell proliferation and inhibits cell apoptosis in Rb, which suggested an oncogenic role of miR-498 in Rb. Meanwhile, we supported the role of CCPG1 that was the direct target of oncogenic miR-498 as the tumor suppressor in Rb. These results shed new light on the role of miR-498 in Rb and may provide a novel strategy for treatment of Rb.
References
American Cancer Society (2003) Neoplasms of the Eye. Cancer Medicine. Hamilton, Ontario: BC Decker Inc 85
Bagnyukova TV, Pogribny IP, Chekhun VF (2006) MicroRNAs in normal and cancer cells: a new class of gene expression regulators. Exp Oncol 28:263–269
Balmer A, Zografos L, Munier F (2006) Diagnosis and current management of retinoblastoma. Oncogene 25:5341–5349
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z (2005) Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 37:766–770
Broaddus E, Topham A, Singh AD (2009) Incidence of retinoblastoma in the USA: 1975–2004. Br J Ophthalmol 93:21–23
Cavenee WK, Dryja TP, Phillips RA, Benedict WF, Godbout R, Gallie BL, Murphree AL, Strong LC, White RL (1983) Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature 305:779–784
Chen D, Livne-Bar I, Vanderluit JL, Slack RS, Agochiya M, Bremner R (2004) Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell 5:539–551
Cong J, Liu R, Wang X, Wang J, Wang H, Hou J (2015) Low miR-498 expression levels are associated with poor prognosis in ovarian cancer. Eur Rev Med Pharmacol Sci 19:4762–4765
Deng S, Calin GA, Croce CM, Coukos G, Zhang L (2008) Mechanisms of microRNA deregulation in human cancer. Cell Cycle 7:2643–2646
Doe JM (2008) MicroRNA: cancer and future treatment applications. Basic Biotech e J 36–42
Du W, Pogoriler J (2006) Retinoblastoma family genes. Oncogene 25:5190–5200
Elkington AR, Khaw PT (1988) ABC of eyes. Squint. BMJ 297:608–611
Esquela-Kerscher A, Slack FJ (2006) Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer 6:259–269
Faghihi MA, Kocerha J, Modarresi F, Engström PG, Chalk AM, Brothers SP, Koesema E, St Laurent G, Wahlestedt C (2010) RNAi screen indicates widespread biological function for human natural antisense transcripts. PLoS One 5:e13177
Hammond SM (2007) MicroRNAs as tumor suppressors. Nat Genet 39:582–583
Hobert O (2008) Gene regulation by transcription factors and microRNAs. Science 319:1785–1786
Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, Trent JM, Staudt LM, Hudson J Jr, Boguski MS, Lashkari D, Shalon D, Botstein D, Brown PO (1999) The transcriptional program in the response of human fibroblasts to serum. Science 283:83–87
Knudson AG Jr (1971) Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A 68:820–823
Liu R, Liu F, Li L, Sun M, Chen K (2015) MiR-498 regulated FOXO3 expression and inhibited the proliferation of human ovarian cancer cells. Biomed Pharmacother 72:52–57
Lohmann DR, Gallie BL (2004) Retinoblastoma: revisiting the model prototype of inherited cancer. Am J Med Genet C Semin Med Genet 129C:23–28
MacCarthy A, Birch JM, Draper GJ, Hungerford JL, Kingston JE, Kroll ME, Onadim Z, Stiller CA, Vincent TJ, Murphy MF (2009) Retinoblastoma in Great Britain 1963–2002. Br J Ophthalmol 93:33–37
MacCarthy A, Draper GJ, Steliarova-Foucher E, Kingston JE (2006) Retinoblastoma incidence and survival in European children (1978–1997). Report from the automated childhood cancer information system project. Eur J Cancer 42:2092–2102
Pfeffer S, Voinnet O (2006) Viruses, microRNAs and cancer. Oncogene 25:6211–6219
Schepeler T, Reinert JT, Ostenfeld MS, Christensen LL, Silahtaroglu AN, Dyrskjot L, Wiuf C, Sørensen FJ, Kruhoffer M, Laurberg S, Kauppinen S, Orntoft TF, Andersen CL (2008) Diagnostic and prognostic MicroRNAs in stage II colon cancer. Cancer Res 68:6416–6424
Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS (2010) Non-coding RNAs: regulators of disease. J Pathol 220:126–139
Ventura A, Jacks T (2009) MicroRNAs and cancer. Short RNAs go a long way. Cell 136:586–591
Wang M, Zhang Q, Wang J, Zhai Y (2015) MicroRNA-498 is downregulated in non-small cell lung cancer and correlates with tumor progression. J Cancer Res Ther 11:C107–C111
Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, Xiao S, Lu H (2008) miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Investig 88:1358–1366
Zhao JJ, Yang J, Lin JH, Yao N, Zhu YH, Zheng JH, JH X, Cheng JQ, Lin JY, Ma X (2009) Identification of miRNAs associated with tumorigenesis of retinoblastoma by miRNA microarray analysis. Childs Nerv Syst 25:13–20
Acknowledgements
We thank Chao Lu from Department of Pediatrics, The First Affiliated Hospital, Nanjing Medical University, Nanjing, China, for his kind help in the current study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None.
Rights and permissions
About this article
Cite this article
Yang, L., Wei, N., Wang, L. et al. miR-498 promotes cell proliferation and inhibits cell apoptosis in retinoblastoma by directly targeting CCPG1. Childs Nerv Syst 34, 417–422 (2018). https://doi.org/10.1007/s00381-017-3622-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00381-017-3622-8