Abstract
In the recent decades, carotid angioplasty and stenting (CAS) has been developed into a credible option for the patients with carotid stenosis. However, restenosis remains a severe and unsolved issue after CAS treatment. Restenosis is characterized by neointimal hyperplasia, which is partially caused by vascular smooth muscle cells (VSMC) proliferation. However, the molecular mechanism involved in the restenosis is still unclear. In this study, we demonstrated a functional crosstalk between two TGF-β superfamily signaling pathway members, Smad3 and BMPR2, in VSMC proliferation. Smad3 plays an important role in the TGF-β/Smad3 signaling pathway, and is significantly upregulated in the carotid artery with restenosis to promote VSMC proliferation. In contrast, BMP receptor II (BMPR2), an inhibitor of VSMC proliferation is downregulated in carotid restenosis. We further found that BMPR2 downregulation is mediated by miR-17–92 cluster, which is transcriptionally regulated by Smad3. Thus, Smad3 upregulation and Smad3/miR-17–92 cluster-dependent BMPR2 downregulation are likely to promote VSMC proliferation and restenosis. Taken together, our results may provide novel clues for early diagnosis of carotid restenosis and developing new therapeutic strategy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Carotid stenosis is a narrowing or constriction of the inner surface of the carotid artery [1, 2]. It is a major risk factor for stroke that leads to brain damage [3, 4]. Carotid stenosis is usually caused by atherosclerosis [5, 6], characterized by the atherosclerostic plagues accumulating in the artery wall, thus occluding the blood flow. In the last decade, carotid angioplasty and stenting (CAS) has been developed into a credible alternative treatment to carotid endarterectomy (CEA) for the patients with symptomatic moderate- and high-grade stenosis [7, 8]. However, restenosis still remains an unsolved issue following CAS treatment, and the restenosis rate is even higher than that following CEA treatment [9, 10]. Restenosis is a healing process of the arterial wall in response to mechanical injury caused by CAS and comprises two major processes—neointimal hyperplasia and vessel remodeling [11, 12]. Thus, to develop more effective therapeutic approaches for preventing restenosis, a better understanding of the molecular mechanism of restenosis is important.
One of the important characters of restenosis is vascular smooth muscle cells (VSMC) proliferation, which is likely to be mediated by transforming growth factor-beta (TGF-β)/bone morphogenic protein (BMP) signaling pathway. TGF-β is known to play an important role in the development of restenosis [13, 14]. As a profibrotic cytokine, TGF-β enhances neointimal formation by stimulating extracellular matrix (ECM) synthesis [15, 16]. Among the many downstream effectors of TGF-β, Smad3 might play a critical role in the formation of intimal hyperplasia. It has been shown that TGF-β1, through Smad3-dependent pathway, stimulates VSMC fibronectin synthesis, thus enhancing neointimal ECM accumulation [17]. Moreover, it has been shown that Smad3 overexpression also enhanced intimal hyperplasia via the stimulation of VSMC proliferation [13, 15]. Smad3 acts as a transcription factor that regulates the expression of a number of genes in the cellular processes [18, 19]. However, how Smad3 regulates in the carotid artery restenosis is largely unknown.
Besides TGF-β, bone morphogenetic protein (BMP) is another important member of the TGF-β superfamily [20, 21]. To date, around 20 BMP family members have been identified and characterized [22]. BMPs signal through serine/threonine kinase receptors, composed of type I and II subtypes [23]. Three type I receptors have been shown to bind BMP ligands, type IA and IB BMP receptors (BMPR1A and BMPR1B), and type IA activin receptor (ActR1A) [23]. Three type II receptors for BMPs have also been identified, including type II BMP receptor (BMPR2), and type II and IIB activin receptors (ActR2 and ActR2B) [23]. The function of BMP-dependent signaling pathway in carotid artery restenosis is not clear. However, loss-of-function mutations of BMP receptor II (BMPR2) are often identified in the patients with familial pulmonary arterial hypertension (PAH) [24, 25], a disease characterized by excessive VSMC proliferation, particularly in response to TGF-β [26]. Thus, BMPR2 is likely to act as a negative regulator to suppress TGF-β-dependent VSMC proliferation. Since VSMC proliferation causes neointimal formation in carotid restenosis [11, 12], we hypothesize that Smad3-dependent signaling pathway has functional crosstalk with BMPR2 in VSMC proliferation during carotid restenosis.
Using human tissue samples, we confirmed that Smad3 is significantly upregulated in carotid restenosis. Moreover, we found that the expression level of BMPR2 was significantly reduced in human carotid artery restenosis. The ChIP analyses showed that BMPR2 was targeted by miR-17–92 cluster members, which is controlled by Smad3. Thus, our results reveal a novel crosstalk between Smad3 and BMPR2 through miR-17–92 cluster in carotid artery restenosis. This study may provide a novel therapeutic strategy for carotid artery restenosis, namely, inhibition of miR-17–92 cluster, which may not only block the Smad3 function but also rescue BMPR2 expression to relieve the neointimal formation.
Materials and methods
Sample collection
The carotid artery tissues were surgically removed from the patients with carotid restenosis following CAS treatment at Beijing Xuanwu Hospital in China, and were harvested for this study in accordance with informed consent principles.
Peripheral blood samples were collected from the patients with carotid restenosis following CAS treatment at Beijing Xuanwu Hospital in China after informed consent from the subjects. Peripheral blood samples from healthy volunteers were also collected as control. All the samples used in this study were harvested after obtaining approval from the ethics committees at Capital Medical University of China and University of Michigan.
Cell culture
Human carotid artery smooth muscle cells (hHCtASMCs) were obtained commercially (Cell Applications, Inc.) and cultured in Medium 231 supplemented with Smooth Muscle Growth Supplement (Life technologies) in 37 °C with 5 % CO2.
Histopathology and immunofluorescent staining
The artery specimens were fixed in 4 % paraformaldehyde for 24 h, dehydrated, and embedded into paraffin wax, then sectioned to 4 μm slides and processed for H&E staining.
To perform immunofluorescent staining, sections were deparaffinized and incubated with rabbit anti-Smad3 (1:500) (Upstate, Cat. # 04-1035) and mouse anti-BMPR2 (1:500) (Millipore, Cat. # MABD171). The slides were then washed in PBS and incubated with FITC-coupled secondary antibodies (1:1000, Jackson ImmunoResearch Laboratories).
Transfection of siRNAs to hHCtASMCs
hHCtASMCs were transfected with 500 ng of Smad3 siRNA and Mock siRNAs using a Primary P1 Nucleofection Kit and 4D Nucleofector machine (Lonza). Smad3 siRNA sequence was ordered from Dharmacon. The siRNA sense sequence is 5′-GAGAAACCAGUGACCACCA-3′, and the mock siRNA sense sequence is 5′-CUACAACUCCCACAACGUA-3′.
RNA isolation and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was isolated from the carotid artery tissue using Trizol (Invitrogen) according to the manufacturer’s instruction. For circulating RNA isolation, the whole blood was allowed to stand overnight at 4 °C, and total circulating RNA was isolated from 200 μl serum using miRNeasy mini kit according to the manufacturer’s protocol (Qiagen).
Approximately, 500 ng of total RNA was reverse transcribed using oligo-dT primers. The cDNA was utilized as a template to amplify target genes with SYBR Premix Ex Tag Kit (TaKaRa). Specific primers were listed in Supplementary Table 1. Each RNA sample was evaluated in triplicate. Gene expression results were analyzed by the ΔΔCt method and normalized to β-actin expression. The qRT-PCR assay was performed on Bio-Rad iQ5 instrument. The data were analyzed using Optical System Software 2.0.
The expression levels of mature miRNAs were determined using miRNA-specific qRT-PCR. The specific RT and PCR primers for miR-17, 18a, 19a, 20a, 92a, 130a, 21, and U6 were purchased from Qiagen. Reverse transcription of miRNAs was performed with a miScript Reverse Transcription Kit (QiAGEN). The qPCR was performed on Bio-Rad IQ 5 instrument by means of SYBR Premix Ex Tag Kit (TaKaRa). The expression levels were normalized to the U6 endogenous control and measured by the comparative Ct (ΔΔCt) method.
Dual luciferase reporter gene construct and dual luciferase reporter assay
The target site of microRNA with ~60 flanking sequence in BMPR2 3′ UTR was synthesized with cleavage sites for Xba I (5′ end) and Not I (3′ end), and a mutated sequence of the binding site was also synthesized. The two constructs were termed as WT and Mut. The DNA oligos were cloned into the pRL-TK vector (Promega). The DNA sequences containing the miR-17–92 cluster, miR-130a, and miR-21 target sites as well as the mutant are shown in Supplementary Table 1. Each vector, along with 100 ng of PGL3 and 200 nmol/L miR-17–92 cluster, miR-130 and miR-21 or mimic control, was transfected into 293T cells. Cells were assayed for renilla and firefly luciferase activity using the Dual-Luciferase Reporter System (Promega).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation assays (ChIP) for artery tissues were performed according to the protocol described by Upstate. In brief, 30 mg of fresh artery tissues per antibody in every ChIP were chopped into small pieces and transferred into 10 mL PBS (plus protease inhibitor). The artery tissues were fixed by 1 % formaldehyde at room temperature for 15 min, and then stopped by adding fresh glycine to a final concentration of 0.125 M at temperature for 5 min. After being washed by cold PBS twice, the artery tissues were resuspended in 1 mL cold PBS (plus protease inhibitors). To achieve a single cell suspension, the Medimachine from Becton–Dickinson was used to grind tissue for 2 min. After centrifugation, the cell pellet was suspended in 6× volume of cell lysis buffer plus protease inhibitors for 15 min on ice, and lysed using a B dounce several times to aid in nuclei release. Cell nuclei were collected by centrifugation at 1,000×g at 4 °C. The cell nuclei were resuspended in 5 × volume of nuclei lysis buffer plus the protease inhibitors, incubated on ice for 20 min and then subjected to sonication. The artery tissue DNA was sonicated to an average size between 300 and 600 bp. Solubilized chromatin was immunoprecipitated with antibody against Smad3 (Upstate), and IgG was used as control. Antibody–chromatin complexes were pulled down using protein A-Sepharose. After crosslink reversal and proteinase K treatment, immunoprecipitated DNA was extracted with phenol–chloroform, ethanol precipitated, and treated with RNase. ChIP DNA was subjected to PCR amplification using primers flanking the Smad3-binding site at the miRNA gene promoter.
Protein extraction and western blotting
Protein samples were extracted from the artery tissues by using total protein extraction kit (Millipore). The protein concentration was determined using BCA standard curve. Equal amounts of protein extracts were separated by SDS-PAGE and transferred electrophoretically to PVDF membranes (Millipore). The membranes were blocked in TBST containing 5 % milk at room temperature for 1 h. After washing with TBST, the blocked membranes were probed with rabbit anti-Smad3 (1:500) (Upstate), mouse anti-BMPR2 (1:500) (Millipore), and mouse anti-β-actin (1:10,000) (Sigma) primary antibodies overnight at 4 °C. After three consecutive 10-min washes with TBST, the membranes were incubated with HRP-conjugated goat-anti-rabbit or goat-anti-mouse secondary antibody for 1 h. The membranes were washed again three times with TBST and developed using the ECL+ detection system (GE Healthcare).
Statistical analysis
All the experiments were performed at least three times. Two-tailed Student’s t test was used for comparing the two groups. ANOVA was used for multiple-group (>2) comparison. Data were presented as mean ± SD p < 0.05 were considered statistically significant.
Result and discussion
In the last decade, CAS has been proposed as a safe and an effective alternative to CEA for patients with carotid stenosis [7, 8]. However, restenosis remains a severe and unsolved issue following CAS treatment [9, 10]. One option for these patients is to surgically remove the blocked part of the carotid artery together with stent, and use graft to replace the removed vessels [27]. We harvested the carotid artery samples following this type of surgery. H&E staining was performed, and neointima hyperplasia was observed in the carotid artery with stenosis, but not in the adjacent normal artery tissue (Fig. 1a).
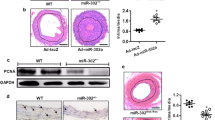
Expression changes of Smad3 and BMPR2 in carotid restenosis. a Carotid artery with restenosis was surgically removed from the patients and harvested for H&E staining. Thickened neointima was observed (dark arrows). Adjacent normal carotid artery was shown as the control. Scale bars 400 μm. b Immunofluorescence staining was performed to examine the expression level of Smad3 and BMPR2 in the carotid artery tissues. Significant increase of Smad3 was observed in the carotid artery with restenosis. In contrast, BMPR2 is downregulated in carotid restenosis. The adjacent normal artery was used as the control. * artery wall; # neointima. Scale bars 200 μm. c The result of qRT-PCR shows the significant increase of Smad3 in carotid artery restenosis. Number 1, 2, 3, 4, 5, 6, and 7 represent the carotid tissue samples from different patients. Error bars indicate SD (n = 6) ** p < 0.01. d Western blotting was used to detect the protein levels of Smad3 and BMPR2. Smad3 upregulation and BMPR2 downregulation were observed in carotid restenosis. β-actin was used as protein-loading control. e Quantitative analysis of the Western blotting results was performed using ImageJ. Around 80 % decrease of BMPR2 was observed in carotid restenosis. Error bars indicate SD (n = 6) ** p < 0.01. f The result of qRT-PCR shows a slight decrease of BMPR2 mRNA level in carotid restenosis. Error bars indicate SD (n = 6) * p < 0.05
TGF-β1/Smad3 signaling pathway has been shown to play a key role for promoting the neointimal formation during carotid restenosis, including VSMC proliferation, through experimental studies in the animal models [15, 16]. Thus, we examined the Smad3 expression levels in human carotid artery tissue with restenosis by immunofluorescence staining. As shown in Fig. 1b, Smad3 was positively stained in the neointima hyperplasia but not normal carotid artery. Moreover, we confirmed the upregulation of Smad3 using both RT-PCR and Western blotting (Fig. 1c, d). Taken together, these results suggest that the expression of Smad3 was significantly increased in the carotid restenosis.
Besides Smad3, BMPR2 also regulates VSMC proliferation. Thus, we questioned whether BMPR2 participated in developing carotid restenosis. Again, we examined the expression of BMPR2 in carotid restenosis. Both the immunofluorescence staining and Western blotting results indicated a significant decrease of the protein level of BMPR2 in carotid tissues with restenosis (Fig. 1b, c). With the quantitative analysis of Western blotting results, we found a ~80 % decrease of BMPR2 in carotid restenosis (Fig. 1e). However, the result of qRT-PCR showed a slight decrease of BMPR2 mRNA level in artery tissues with restenosis compared with that in normal adjacent artery tissues (Fig. 1f).
This inconsistent alteration of the expression of BMPR2 in mRNA and protein levels indicated that the downregulation of BMPR2 could be mediated by microRNAs, which post-transcriptionally downregulated the target genes [28]. Thus, we then used online software, TargetScan, to predict the candidate microRNAs that potentially regulate the expression of BMPR2 [29]. Several microRNAs were shown to potentially target the 3′ UTR of BMPR2 (Fig. 2a, b). Interestingly, four of these candidate microRNAs, including miR-17, miR-19ab, miR-20a, and miR-92a, belong to miR-17–92 cluster. Conserved target sites of these four microRNAs were found in human BMPR2 3′ UTR (Fig. 2a). Of note, miR-17 and miR-20a share the same potential target sites, because these two microRNAs have the same seed sequence in the 5′ ends (Fig. 2a). In addition, miR-130a and miR-21 are also found to potentially target BMPR2 among prediction results (Fig. 2b). Luciferase assays were performed to examine whether BMPR2 was directly regulated by these six microRNAs. The target site of each of these six microRNAs in BMPR2 3′ UTR was cloned into the downstream of renilla luciferase coding frame of pRL-TK vector (Promeg). Each microRNA targeting site mutant was also generated. The result showed that luciferase signal could be significantly downregulated by miR-17, miR-19a, miR-20a, or miR-92a, but not by miR-130a or miR-21 (Fig. 2c). This result indicated that it is miR-17–92 cluster members, but not miR-130a and miR-21, directly targeting BMPR2. Both miR-130a and miR-21 have been shown to be involved in the carotid restenosis by targeting Gax and PTEN, respectively [30–33]. Both Gax and PTEN can inhibit the proliferation of VSMC [34, 35]. Thus, miR-130a and miR-21 are upregulated in the carotid restenosis and are likely to promote VSMC proliferation by downregulating the expression levels of Gax and PTEN, respectively.
BMPR2 is regulated by miR-17–92 cluster. a Targeting sites of four miR-17–92 cluster members in BMPR2 3′ UTR were predicted by TargetScan. b Targeting sites of miR-130a and miR-21 in BMPR2 3′ UTR were predicted by TargetScan. c Luciferase assays were performed to confirm that BMPR2 was the target gene of miR-17, miR-19a, miR-20a, and miR-92a, but not that of miR-130a and miR-21. Error bars indicate SD (n = 6) ** p < 0.01. d The result of qRT-PCR shows that miR-17–92 cluster members were significantly upregulated in carotid artery restenosis. Error bars indicate SD (n = 7) ** p < 0.01. e Circulating miR-17–92 cluster members increased in sera of the patients with carotid restenosis. Circulating RNAs were isolated from eight patients’ sera, and qRT-PCR was performed to detect the microRNA expression levels. The circulating microRNAs in the healthy volunteers’ sera were detected as the control. ** p < 0.01
We next detected the expression level of miR-17, miR-19a, miR-20a, miR-92a as well as another miR-17–92 family member, miR-18a in the carotid artery tissues with restenosis. All these microRNAs were found to be significantly upregulated in carotid restenosis compared with that in adjacent normal artery (Fig. 2d). These results further support that miR-17–92 cluster members induce the carotid restenosis formation by suppressing the expression of BMPR2. Of note, as a member of miR-17–92 cluster, miR-18a was also upregulated in carotid artery restenosis although little evidence indicate that miR-18a is involved in artery restenosis. It is possible that the upregulation of miR-18a might be a byproduct of expressing miR-17–92 cluster in the carotid artery tissue with restenosis (Fig. 2d).
Recently, extracellular miRNAs have been found to circulate in the bloodstream, and the circulating miRNAs are remarkably stable [36–38]. Although the precise cellular release mechanism of miRNAs remains elusive, some studies revealed that these circulating miRNAs can be delivered to recipient cells, where they can regulate the translation of target genes [39, 40]. It raises the possibility that miRNAs can be probed in the blood and can serve as novel diagnostic markers [38, 41]. Thus, as a potential clinical application for the early diagnosis of the restenosis, we examined the serum levels of four circulating miR-17–92 cluster members. We isolated the serum microRNAs from eight patients with carotid restenosis following CAS as well as eight healthy volunteers. The qRT-PCR was performed to detect the circulating microRNA levels. The upregulation of miR-17–92 cluster members, including miR-17, miR-19a, miR-20a, and miR-92a, was clearly observed in the sera from the patients (Fig. 2e).
Two TGF-β superfamily signaling pathways were involved in the process of human carotid artery restenosis, in which Smad3 is upregulated, and BMPR2 was downregulated. We next asked whether these two signaling pathways have crosstalk in carotid restenosis. Smad3 is one of the core transcription factors of TGF-β signaling pathway, and has been shown to directly bind to the promoter of miR-200b/a as a transcription activator in gastric cancer cells [42]. The Smad-binding elements have also been identified, and the consensus sequence is GTCTG [42]. Here, we hypothesize that Smad3 might regulates BMPR2 via the activation of miR-17–92 cluster during restenosis. We found that the Smad-binding elements existed in the promoter region of miR-17–92 cluster (Fig. 3a). ChIP–qRT-PCR was then performed, and we found that Smad3 indeed stayed in the promoter region of miR-17–92 gene cluster but not in the promoter region of miR-20b and miR-92b (Fig. 3b). To demonstrate that Smad3 activates the transcription of miR-17–92 gene cluster and suppresses the expression of BMPR2, we knocked down Smad3 in human carotid artery smooth muscle cells by siRNA (Fig. 3c). Compared with control siRNA treatment, loss of Smad3 significantly suppressed the expression of miR-17–92 gene cluster (Fig. 3d). Moreover, lacking Smad3 induced the upregulation of the expression level of BMPR2 (Fig. 3e).
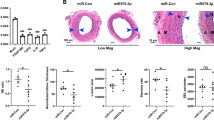
miR-17–92 cluster is regulated by Smad3. a Smad-binding sequence, GTCTG, was found in the promoter region of miR-17–92 gene cluster. b ChIP-qPCR results suggested that the promoter region of miR-17–92 gene cluster was bound by Smad3. Carotid artery tissues with restenosis from 7 patients were used for ChIP-qPCR assay. An irrelevant IgG was used as control. Error bars indicate SD (n = 7) ** p < 0.01 versus IgG. c hHCtASMCs were transfected with Smad3 and Mock siRNA respectively. And qRT-PCR was performed to detect the Smad3 mRNA level in hHCtASMCs. Smad3 siRNA can effectively decrease the Smad3 mRNA level. Error bars indicate SD (n = 6) ** p < 0.01 versus Mock. d Knockdown Smad3 suppresses the transcription of miR-17–92 gene cluster. hHCtASMCs were treated with Smad3 siRNA. The qRT-PCR was performed to detect the microRNA expression level. Error bars indicate SD (n = 6) ** p < 0.01. e Western blotting was performed to examine the protein level of BMPR2. Significant increase of BMPR2 was observed in hHCtASMCs treated with Smad3 siRNA but not in the cells treated with Mock siRNA. β-actin was used as protein loading control
Taken together, our study demonstrates that miR-17–92 cluster mediates the crosstalk between the two important TGF-β superfamily signaling pathways, TGF-β1/Smad3 and BMP/BMPR2, to promote the VSMC proliferation and neointimal formation in the carotid artery restenosis (Fig. 4).
A model shows the functional crosstalk between Smad3 and BMPR2 via miR-17–92 in carotid artery restenosis. Smad3 is significantly upregulated in the carotid artery with restenosis to promote VSMC proliferation. Upregulation of Smad3 activates the transcription of miR-17–92 cluster. Then, miR-17–92 family members downregulate BMPR2, an inhibitor of VSMC proliferation, to further promote VSMC proliferation and carotid artery restenosis
References
Meissner I, Meyer FB (1994) Carotid stenosis and carotid endarterectomy. Cerebrovasc Brain Metab Rev 6(2):163–179
Berman SS, Bernhard VM, Erly WK, McIntyre KE, Erdoes LS, Hunter GC (1994) Critical carotid artery stenosis: diagnosis, timing of surgery, and outcome. J Vasc Surg 20(4):499–508 Discussion 508–410
Kelly R (1992) Selections from current literature: prevention of stroke in non-rheumatic atrial fibrillation and carotid artery stenosis. Fam Pract 9(2):231–236
Sherman DG (1989) The carotid artery and stroke. Am Fam Physician 40 (5 Suppl):41S–44S, 47S–49S
Parnetti L, Mercuri M, Susta A, Lupattelli G, Ciuffetti G, Senin U (1988) Extracranial carotid atherosclerosis evaluation and stroke occurrence: role of the echotomographic analysis. Angiology 39(8):705–713
Wasserman BA, Haacke EM, Li D (1994) Carotid plaque formation and its evaluation with angiography, ultrasound, and MR angiography. J Magn Reson Imaging 4(4):515–527
Fanelli F, Boatta E, Cannavale A, Corona M, Lucatelli P, Wlderk A, Cirelli C, Salvatori FM (2012) Carotid artery stenting: analysis of a 12-year single-center experience. J Endovasc Ther 19(6):749–756. doi:10.1583/JEVT-12-3944MR.1
Gahremanpour A, Perin EC, Silva G (2012) Carotid artery stenting versus endarterectomy: a systematic review. Tex Heart Inst J 39(4):474–487
AbuRahma AF, Abu-Halimah S, Hass SM, Nanjundappa A, Stone PA, Mousa A, Lough E, Dean LS (2010) Carotid artery stenting outcomes are equivalent to carotid endarterectomy outcomes for patients with post-carotid endarterectomy stenosis. J Vasc Surg 52(5):1180–1187. doi:10.1016/j.jvs.2010.06.074
Cosottini M, Michelassi MC, Bencivelli W, Lazzarotti G, Picchietti S, Orlandi G, Parenti G, Puglioli M (2010) In stent restenosis predictors after carotid artery stenting. Stroke Res Treat. doi:10.4061/2010/864724
Costa MA, Simon DI (2005) Molecular basis of restenosis and drug-eluting stents. Circulation 111(17):2257–2273. doi:10.1161/01.CIR.0000163587.36485.A7
Inoue T, Node K (2009) Molecular basis of restenosis and novel issues of drug-eluting stents. Circ J 73(4):615–621
Suwanabol PA, Seedial SM, Shi X, Zhang F, Yamanouchi D, Roenneburg D, Liu B, Kent KC (2012) Transforming growth factor-beta increases vascular smooth muscle cell proliferation through the Smad3 and extracellular signal-regulated kinase mitogen-activated protein kinases pathways. J Vasc Surg 56(2):446–454. doi:10.1016/j.jvs.2011.12.038
Suwanabol PA, Kent KC, Liu B (2011) TGF-beta and restenosis revisited: a Smad link. J Surg Res 167(2):287–297. doi:10.1016/j.jss.2010.12.020
Tsai S, Hollenbeck ST, Ryer EJ, Edlin R, Yamanouchi D, Kundi R, Wang C, Liu B, Kent KC (2009) TGF-beta through Smad3 signaling stimulates vascular smooth muscle cell proliferation and neointimal formation. Am J Physiol Heart Circ Physiol 297(2):H540–H549. doi:10.1152/ajpheart.91478.2007
Rasmussen LM, Wolf YG, Ruoslahti E (1995) Vascular smooth muscle cells from injured rat aortas display elevated matrix production associated with transforming growth factor-beta activity. Am J Pathol 147(4):1041–1048
Ryer EJ, Hom RP, Sakakibara K, Nakayama KI, Nakayama K, Faries PL, Liu B, Kent KC (2006) PKCdelta is necessary for Smad3 expression and transforming growth factor beta-induced fibronectin synthesis in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol 26(4):780–786. doi:10.1161/01.ATV.0000209517.00220.cd
Massague J, Seoane J, Wotton D (2005) Smad transcription factors. Genes Dev 19(23):2783–2810. doi:10.1101/gad.1350705
Derynck R, Zhang Y, Feng XH (1998) Smads: transcriptional activators of TGF-beta responses. Cell 95(6):737–740
Matzuk MM (1995) Functional analysis of mammalian members of the transforming growth factor-beta superfamily. Trends Endocrinol Metab 6(4):120–127
Miyazono K, Kusanagi K, Inoue H (2001) Divergence and convergence of TGF-beta/BMP signaling. J Cell Physiol 187(3):265–276. doi:10.1002/jcp.1080
Chen D, Zhao M, Mundy GR (2004) Bone morphogenetic proteins. Growth Factors 22(4):233–241. doi:10.1080/08977190412331279890
Sieber C, Kopf J, Hiepen C, Knaus P (2009) Recent advances in BMP receptor signaling. Cytokine Growth Factor Rev 20(5–6):343–355. doi:10.1016/j.cytogfr.2009.10.007
Cogan J, Austin E, Hedges L, Womack B, West J, Loyd J, Hamid R (2012) Role of BMPR2 alternative splicing in heritable pulmonary arterial hypertension penetrance. Circulation 126(15):1907–1916. doi:10.1161/CIRCULATIONAHA.112.106245
Li W, Dunmore BJ, Morrell NW (2010) Bone morphogenetic protein type II receptor mutations causing protein misfolding in heritable pulmonary arterial hypertension. Proc Am Thorac Soc 7(6):395–398. doi:10.1513/pats.201002-024AW
Nasim MT, Ogo T, Chowdhury HM, Zhao L, Chen CN, Rhodes C, Trembath RC (2012) BMPR-II deficiency elicits pro-proliferative and anti-apoptotic responses through the activation of TGFbeta-TAK1-MAPK pathways in PAH. Hum Mol Genet 21(11):2548–2558. doi:10.1093/hmg/dds073
Jost D, Unmuth SJ, Meissner H, Henn-Beilharz A, Henkes H, Hupp T (2012) Surgical treatment of carotid in-stent-restenosis: novel strategy and current management. Thorac Cardiovasc Surg 60(8):517–524. doi:10.1055/s-0032-1311535
Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9(2):102–114. doi:10.1038/nrg2290
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120(1):15–20. doi:10.1016/j.cell.2004.12.035
Chen Y, Gorski DH (2008) Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood 111(3):1217–1226. doi:10.1182/blood-2007-07-104133
Wu WH, Hu CP, Chen XP, Zhang WF, Li XW, Xiong XM, Li YJ (2011) MicroRNA-130a mediates proliferation of vascular smooth muscle cells in hypertension. Am J Hypertens 24(10):1087–1093. doi:10.1038/ajh.2011.116
Maegdefessel L, Azuma J, Toh R, Deng A, Merk, Raiesdana A, Leeper NJ, Raaz U, Schoelmerich AM, McConnell MV, Dalman RL, Spin JM, Tsao PS (2012) MicroRNA-21 blocks abdominal aortic aneurysm development and nicotine-augmented expansion. Sci Transl Med 4(122):122ra122. doi:10.1126/scitranslmed.3003441
McDonald RA, White KM, Wu J, Cooley BC, Robertson KE, Halliday CA, McClure JD, Francis S, Lu R, Kennedy S, George SJ, Wan S, van Rooij E, Baker AH (2013) miRNA-21 is dysregulated in response to vein grafting in multiple models and genetic ablation in mice attenuates neointima formation. Eur Heart J 34(22):1636–1643. doi:10.1093/eurheartj/eht105
Perlman H, Luo Z, Krasinski K, Le Roux A, Mahfoudi A, Smith RC, Branellec D, Walsh K (1999) Adenovirus-mediated delivery of the Gax transcription factor to rat carotid arteries inhibits smooth muscle proliferation and induces apoptosis. Gene Ther 6(5):758–763. doi:10.1038/sj.gt.3300893
Furgeson SB, Simpson PA, Park I, Vanputten V, Horita H, Kontos CD, Nemenoff RA, Weiser-Evans MC (2010) Inactivation of the tumour suppressor, PTEN, in smooth muscle promotes a pro-inflammatory phenotype and enhances neointima formation. Cardiovasc Res 86(2):274–282. doi:10.1093/cvr/cvp425
Kroh EM, Parkin RK, Mitchell PS, Tewari M (2010) Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 50(4):298–301. doi:10.1016/j.ymeth.2010.01.032
Kosaka N, Iguchi H, Ochiya T (2010) Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 101(10):2087–2092. doi:10.1111/j.1349-7006.2010.01650.x
Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 105(30):10513–10518. doi:10.1073/pnas.0804549105
Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, Sun F, Lu J, Yin Y, Cai X, Sun Q, Wang K, Ba Y, Wang Q, Wang D, Yang J, Liu P, Xu T, Yan Q, Zhang J, Zen K, Zhang CY (2010) Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell 39(1):133–144. doi:10.1016/j.molcel.2010.06.010
Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT (2011) MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 13(4):423–433. doi:10.1038/ncb2210
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Zen K, Zhang CY (2008) Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18(10):997–1006. doi:10.1038/cr.2008.282
Ahn SM, Cha JY, Kim J, Kim D, Trang HT, Kim YM, Cho YH, Park D, Hong S (2012) Smad3 regulates E-cadherin via miRNA-200 pathway. Oncogene 31(25):3051–3059. doi:10.1038/onc.2011.484
Acknowledgments
This study was supported by the National Institute of Health (CA132755 and CA130899 to X.Y.), the University of Michigan Cancer Center and GI Peptide Research Center. X.Y. is a recipient of the Era of Hope Scholar Award from the Department of Defense.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Tao Luo and Shijun Cui have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Luo, T., Cui, S., Bian, C. et al. Crosstalk between TGF-β/Smad3 and BMP/BMPR2 signaling pathways via miR-17–92 cluster in carotid artery restenosis. Mol Cell Biochem 389, 169–176 (2014). https://doi.org/10.1007/s11010-013-1938-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-013-1938-6