Abstract
Insertion/deletion (I/D) polymorphism, of a 287-bp Alu repetitive sequence in intron 16 of the angiotensin-converting enzyme (ACE) gene has been shown to be associated with different types of diseases and has been widely investigated in different populations with different ethnic origins. Various reports were published suggesting inter-ethnic variations in the frequency of allelic forms of the ACE gene. The goal of this study was to test the distribution of alleles and the different genotypes of ACE (I/D) polymorphism in Bahraini subjects and compare the results with those obtained from other population studies. The Bahraini population is an Arabic peninsula population with a high prevalence of T2DM and hypertension. A total of 560 unrelated Bahraini individuals were recruited in this study and the presence (insertion)/absence (deletion) (I/D) polymorphism of a 287-bp Alu1 element inside intron 16 of the ACE gene was done by PCR-based assays and the presence or absence of the genotypes were analyzed by the gel electrophoresis. The distribution of II, ID, and DD genotypes showed differences among Bahraini subjects, and the frequency of the D allele was significantly (P < 0.05) higher in the studied group. The results obtained for the D allele are consistent with those obtained from previous studies among Arabs, Africans, and Caucasians, but differs significantly (P < 0.05) from those in Japanese and Chinese, thus proving the ethnic variation in the distribution of the ACE alleles in different populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The angiotensin-converting enzyme (ACE) is a key enzyme of the renin–angiotensin system (RAS) that has long been recognized as crucial in the regulation of systemic blood pressure and renal electrolyte homeostasis [1, 2]. This enzyme acts by converting the inactive Angiotensin I to the vasoconstrictor Angiotensin II, and inactivates the vasodilator proinflammatory bradykinin and substance P peptides [3]. The ACE gene (encoding enzyme type kinase II, EC 3.4.15.1) is located on chromosome 17 and the gene polymorphisms have been widely investigated. The insertion/deletion (I/D) polymorphism of ACE has been discovered and described by Rigat et al. [4]. The polymorphism is characterized by either the presence/insertion (I) or absence/deletion (D) of a 287-base pair Alu1 element inside intron 16 of the ACE gene, producing three genotypes (DD, II homozygote, and ID heterozygote) [4, 5].
Several investigations have been shown that the D allele of an insertion/deletion (I/D) polymorphism of the gene encoding ACE is associated with higher plasma ACE concentrations [4, 5] and suggested an association of the ACE I/D polymorphism with incidence, pathogenesis and progression of several diseases including diabetes mellitus and its long-term macro- and microvascular complications [6–8], including diabetic nephropathy [9–11]. Moreover, other studies have also provide a positive association of the ACE I/D polymorphism and cardiomyopathy [12, 13] and hypertension [14]. Since the ACE I/D polymorphism is also associated with the overall plasma ACE concentration [15], it was suggested that ACE might be a good candidate gene for the metabolic syndrome, which is clustered together with hypertension, DM and dyslipidemia [16]. However, conflicting results have been reported regarding the association between ACE I/D polymorphism and diseases [17–24]. Moreover, various published reports have suggested inter-ethnic variations in the frequency of allelic forms of the ACE genes [25–27].
The aim of our study was to test the distribution of allele and genotype frequencies of ACE (I/D) polymorphism among Bahrainis and to compare the results with those of other ethnic groups. The Bahraini population is an Arabic peninsula population with a high prevalence of T2DM occurring in 20–30% of adult population [28] and hypertension [29]. The characteristics of Arabic population make them ideal for the study of complex, polygenic, multifactorial disorders such as diabetes. Arabic populations are characterized by genetic homogeneity due to tribal structure of their society, large family size and extensive consanguinity, as a result of these factors, the statistical power of susceptibility and target gene discovery studies is greatly increased if homogeneous and consanguineous populations like the Arabic populations are used.
Materials and methods
Study population
This epidemiological study comprised a total of randomly selected 560 unrelated adults Bahrainis attending the two major hospitals in Bahrain: Salmaniya Medical Center (SMC) and Bahrain Defense Force (BDF) with or without problems. Each subject received a questionnaire which included an informed consent form to sign agreeing to participate in the study. The project also had the relevant ethical clearance from the Ethics Committee from the Arabian Gulf University to undertake the study.
DNA extraction
Venous blood was collected by venipuncture from each individual in vacutainer tubes with EDTA as anticoagulant. Peripheral blood mononuclear cells were used as a source of genomic DNA. Extraction of DNA was performed according to the protocol of the QIAamp DNA Blood mini-spin column (QiagenGmbH, D-40724 Hilden, Germany).
ACE I/D polymorphism genotyping
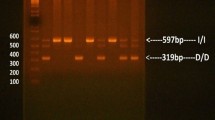
Genotyping was performed for the insertion/deletion (I/D) polymorphisms in intron 16 of ACE gene by PCR-gel electrophoresis on the extracted DNA using a pair of oligonucleotide primers 5′-CTGGAGAC CACTCCCAT CCTTTCT-3′ and 5′-GATGTG GCCATCACATTCG TCAGAT-3′ (Biobasic, Markham, Canada). The reactions were run with 5 pmol of each primer in combination with Taq PCR Master Mix (Qiagen, Valencia, CA). DNA was amplified using cycling conditions described previously [15]. PCR products were separated on 2% agarose gels after staining with Vistra Green TM. Two DNA fragments were detected; 190 bp indicating the deletion allele (D) and 490 bp indicating the insertion (I) allele. Under some conditions, the ACE D allele amplifies more effectively than the longer I allele, resulting in mistyping of the ID as the DD genotype. To eliminate the possibility, each sample that had the DD genotype was re-amplified with an insertion-specific primer pair, which recognized an insertion-specific sequence DD F 5′-TGG GAC CAC AGC GCC CGC CAC TAC-3′ DD R 5′-TCG CCA GCC CTC CCA TGC CCA TAA-3′ [30]. The interpretation of PCR results was based on the detection of a PCR product of a 335 bp band of insertion (I alleles) to determine whether mistype of the DNA, was present, or whether no PCR product appeared on the gel [31].
Data analysis
Statistical analysis was performed using SPSS version 14.0 statistical package for Windows. Allele and genotype frequencies were determined by allele counting and the χ2 (Chi-squared) were used to compare observed versus expected outcomes. A value of P < 0.05 was taken as significant.
Results
Characteristics of the randomly selected Bahraini population
In this study, the Bahraini subjects (n = 560) comprised of 295 (52.6%) males and 265 (47.2%) females with a mean age (±SD) of 49.5 ± 15 years and a mean body mass index (BMI) of 26.34 ± 6.5 kg/m2 (Table 1). In this population, 65 (11.6%) were diagnosed as T2DM based on 1998 World Health Organization diagnostic and classification criteria [32]. In addition, there were 6.2% with Nephropathy, 2.3% with Retinopathy, 1.8% with Neuropathy, 30.3% with Hypertension, 25.3% with Hypercholesterolemia, and 7.3% with Cardiovascular Diseases (Table 1).
Genotypes and allele frequencies of I/D polymorphism of the ACE gene in the randomly selected Bahraini population
Allele distribution of ACE I/D polymorphism among the Bahraini subjects was calculated by allele counting using the Hardy–Weinberg (H–W) equation. The results are summarized in Table 2. The frequency of the I allele was 0.38 while that of the D allele was 0.62. Among the 560 Bahraini individuals, 3 of total 228 of DD genotypes were confirmed to be ID and the frequencies of the II, ID, and DD genotypes of the ACE gene in the studied subjects were 15.5, 44.3, and 40.2%, respectively (Table 2). The gene frequency distribution did not deviate from the H–W equilibrium (p 2 = 0.1444; 2pq = 0.4712; q 2 = 0.3844) (Table 2).
Discussion
The present study investigated for the first time, the frequency of the ACE gene I/D polymorphism among randomly selected unrelated Bahraini subjects and the results obtained were compared with other geographic groups (Table 3). Among the randomly selected unrelated Bahraini subjects, the frequency of the D allele of the ACE I/D gene polymorphism was found to be (0.62), which is consistent with those obtained from the Gulf Region with similar identity to the Omanis (0.71) [33, 34], the Emiratis (0.66) [34, 35], and similar to other Arabs, including such as the Tunisians (0.76) [36], Algerians (0.73) [36], Somalis (0.73) [33, 34], Egyptians from Ismailia (0.68) and from Sinai (0.66) [37], Jordanians (0.66), Sudanese (0.64) [33, 34], and Syrians (0.60) [37].
This high frequency of the D allele among Bahrainis is similar to that observed previously not only among Arabs but also in Africans (0.70–0.60) [25, 38]. This is close to the frequency found in Caucasians (0.46–0.58) [5, 39, 40] but much higher than that found in Japanese (0.33–0.35) [41–43] and Chinese (0.29) [44]. On the other hand, the Yanomami Indians and Samoans have the lowest frequencies, 0.15 and 0.09, respectively [25] (Table 3).
The high level of D allele among Bahrainis might have an impact on the high prevalence of diabetes and its complications in addition to hypertension in Bahraini population, which is one of the highest populations in the world regarding rate of diabetes. The positive association of DD genotype with the development of hypertension was previously reported [7, 14, 45, 46] and with the development of cardiovascular disease [5, 6, 12, 13, 47].
Conclusion
The results obtained for the D allele of the ACE I/D gene polymorphism among Bahrainis is consistent with those obtained from previous studies in Arabs, Africans, and Caucasians. This study adds to the data indicating the wide variations in the distribution of the ACE alleles in different populations and highlights that great care needs to be taken when interpreting clinical data on the association of the ACE alleles with different diseases. Further studies need to be done on diabetic and hypertensive patients among Bahraini population compared with control to study the association of ACE polymorphisms with both diseases.
References
Ehlers MR, Riordan JF (1989) Angiotensin-converting enzyme: new concepts concerning its biological role. Biochemistry 28(13):5311–5318
Wang JG, Staessen JA (2000) Genetic polymorphisms in the renin-angiotensin system: relevance for susceptibility to cardiovascular disease. Eur J Pharmacol 410:289–302
Skidgel RA, Erdös EG (1993) Biochemistry of angiotensin converting enzyme. In: Robertson JIS, Nicholls MG (eds) The renin-angiotensin system, vol 1. Gower Medical Publishers, London, pp 10.10–10.11
Rigat B, Hubert C, Alhenc-Gelas F, Corvol P, Soubrier F (1990) An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene account for half the variance of serum enzyme levels. J Clin Invest 86:1343–1346
Tiret L, Rigat B, Visvikis S, Breda C, Corvol P, Cambien F, Soubrier F (1992) Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am J Hum Genet 51(1):197–205
Kennon B, Petrie JR, Small M, Connell JM (1999) Angiotensin-converting enzyme gene and diabetes mellitus. Diabet Med 16(6):448–458
Ramachandran V, Ismail P, Stanslas J, Shamsudin N, Moin S, Jas M (2008) Association of insertion/deletion polymorphism of angiotensin-converting enzyme gene with essential hypertension and type 2 diabetes mellitus in Malaysian subjects. J Renin Angiotensin Aldosterone Syst 9(4):208–214
Baroudi T, Bouhaha R, Moran-Moguel C, Sanchez-Corona J, Ben Maiz H, KammounAbid H, Benammar-Elgaaied A (2009) Association of the insertion/deletion polymorphism of the angiotensin-converting enzyme gene with type 2 diabetes in two ethnic groups of Jerba Island in Tunisia. J Renin Angiotensin Aldosterone Syst 10(1):35–40
Fava S, Azzopardi J, Ellard S, Hattersley AT (2001) ACE gene polymorphism as a prognostic indicator in patients with type 2 diabetes and established renal disease. Diabetes Care 24:2115–2120
ArzuErgen H, Hatemi H, Agachan B, Camlica H, Isbir T (2004) Angiotensin-I converting enzyme gene polymorphism in Turkish type 2 diabetic patients. Exp Mol Med 36(4):345–350
Palomo-Piñón S, Gutiérrez-Rodríguez ME, Díaz-Flores M, Sánchez-Barrera R, Valladares-Salgado A, Utrera-Barillas D, Durán-Reyes G, Galván-Duarte RE, Trinidad-Ramos P, Cruz M (2009) DD genotype of angiotensin-converting enzyme in type 2 diabetes mellitus with renal disease in Mexican Mestizos. Nephrology (Carlton) 14(2):235–239
Sobti RC, Maithil N, Thakur H, Sharma Y, Talwar KK (2010) Association of ACE and FACTOR VII gene variability with the risk of coronary heart disease in north Indian population. Mol Cell Biochem 341(1–2):87–98
Thameem F, Voruganti VS, He X, Nath SD, Blangero J, MacCluer JW, Comuzzie AG, Abboud HE, Arar NH (2008) Genetic variants in the renin-angiotensin system genes are associated with cardiovascular-renal-related risk factors in Mexican Americans. Hum Genet 124(5):557–559
Bawazier LA, Sja’bani M, Haryana SM, Soesatyo MH, Sadewa AH (2010) Relationship of angiotensin converting enzyme gene polymorphism and hypertension in Yogyakarta, Indonesia. Acta Med Indones 42(4):192–198
Rigat B, Hubert C, Corvol P, Soubrier F (1999) PCR detection of the insertion/deletion polymorphism of the human angiotensin converting enzyme gene (DCP1) (dipeptidylcarboxypeptidase 1). Nucleic Acids Res 20(6):1433
Thomas GN, Tomlinson B, Chan JC, Sanderson JE, Cockram CS, Critchley JA (2001) Renin-angiotensin system gene polymorphisms, blood pressure, dyslipidemia, and diabetes in Hong Kong Chinese: a significant association of the ACE insertion/deletion polymorphism with type 2 diabetes. Diabetes Care 24(2):356–361
Jeunemaitre X, Lifton RP, Hunt SC, Williams RR, Lalouel JM (1992) Absence of linkage between the angiotensin converting enzyme locus and human essential hypertension. Nat Genet 1:72–75
Marre M, Bernadet P, Gallois F, Savagner Y, Guyene TT, Hallab M, Cambien F, Passa P, Alhenc-Gelas F (1994) Relationships between angiotensin I converting enzyme gene polymorphism, plasma levels, and diabetic retinal and renal complications. Diabetes 43(3):384–388
Staessen JA, Wang JG, Ginocchio V, Petrov G, Saavedra AP, Soubrier F, Vlietinck R, Fagard R (1997) The deletion/insertion polymorphism of the angiotensin converting enzyme gene and cardiovascular-renal risk. J Hypertens 15(12, Pt 2):1579–1592
Kunz R, Bork JP, Fritsche L, Ringel J, Sharma AM (1998) Association between the angiotensin-converting enzyme-insertion/deletion polymorphism and diabetic nephropathy: a methodologic appraisal and systematic review. J Am Soc Nephrol 9(9):1653–1663
Taal MW (2000) Angiotensin-converting enzyme gene polymorphisms in renal disease: clinically relevant? Curr Opin Nephrol Hypertens 9(6):651–657
Thomas GN, Young RP, Tomlinson B, Woo KS, Sanderson JE, Critchley JA (2000) Renin-angiotensin-aldosterone system gene polymorphisms and hypertension in Hong Kong Chinese. Clin Exp Hypertens 22(1):87–97
Grimm R, Rettig R (2002) Association studies between the angiotensin-converting enzyme insertion/deletion polymorphism and hypertension: still interesting? J Hypertens 20(6):1049–1051
Tascilar N, Dursun A, Ankarali H, Mungan G, Ekem S, Baris S (2009) Angiotensin-converting enzyme insertion/deletion polymorphism has no effect on the risk of atherosclerotic stroke or hypertension. J Neurol Sci 285(1–2):137–141
Barley J, Blackwood A, Carter ND, Crews DE, Cruickshank JK, Jeffery S, Ogunlesi AO, Sagnella GA (1994) Angiotensin converting enzyme insertion/deletion polymorphism: association with ethnic origin. J Hypertens 12(8):955–957
Altshuler D, Kruglyak L, Lander E (1998) Genetic polymorphisms and disease. N Engl J Med 338(22):1626
Pasha MA, Khan AP, Kumar R, Ram RB, Grover SK, Srivastava KK, Selvamurthy W, Brahmachari SK (2002) Variations in angiotensin-converting enzyme gene insertion/deletion polymorphism in Indian populations of different ethnic origins. J Biosci 27(1 Suppl 1):67–70
Al-Mahroos F, McKeigue PM (1998) High prevalence of diabetes in Bahrainis. Association with ethnicity and raised plasma cholesterol. Diabetes Care 21:936–942
Al-Mahroos F, Al-Roomi K, Mckeigue PM (2000) Relation of high blood pressure to glucose intolerance, plasma lipids and educational status in an Arabian Gulf population. Int J Epidemiol 29:71–76
Shanmugan V, Sell KW, Saha BK (1993) Mistyping ACE heterozygotes. PCR Methods Appl 3(2):120–121
Lindpaintner K, Pfeffer MA, Kreutz R, Stampfer MJ, Grodstein F, LaMotte F, Buring J, Hennekens CH (1995) A prospective evaluation of an angiotensin-converting-enzyme gene polymorphism and the risk of ischemic heart disease. N Engl J Med 332(11):706–711
World Health Organisation (1999) Definition, diagnosis and classification of diabetes mellitus and its complications; Report of a WHO Consultation. Part 1: Diagnosis and classification of diabetes mellitus. Geneva, Switzerland
Al-Hinai AT, Hassan MO, Simsek M, Al-Barwani H, Bayoumi R (2002) Genotypes and allele frequencies of angiotensin converting enzyme (ACE) insertion/deletion polymorphism among Omanis. SQU J Sci Res: Med Sci 4(1–2):25–27
Bayoumi R, Simsek M, Yahya TM, Bendict S, Al-Hinai A, Al-Barwani H, Hassan MO (2006) Insertion/deletion polymorphism in the angiotensin converting enzyme (ACE) gene among Sudanese, Somalis, Emiratis, and Omanis. Hum Biol 78(1):103–108
Frossard PM, Obineche EN, Elshahat YI, Lestringant GG (1997) Deletion polymorphism in the angiotensin-converting enzyme gene is not associated with hypertension in a Gulf Arab population. Clin Genet 51(3):211–213
Comas D, Calafell F, Benchemsi N, Helal A, Lefranc G, Stoneking M, Batzer MA, Bertranpetit J, Sajantila A (2000) Alu insertion polymorphisms in NW Africa and the Iberian Peninsula: evidence for a strong genetic boundary through the Gibraltar Straits. Hum Genet 107:312–319
Salem AH, Batzer MA (2009) High frequency of the D allele of the angiotensin-converting enzyme gene in Arabic populations. BMC Res Notes 8(2):99–104
Rutledge DR, Browe CS, Ross EA (1994) Frequencies of the angiotensinogen gene and angiotensin I converting enzyme (ACE) gene polymorphisms in African Americans. Biochem Mol Biol Int 34(6):1271–1275
Cambien F, Poirier O, Lecerf L, Evans A, Cambou JP, Arveiler D, Luc G, Bard JM, Bara L, Ricard S et al (1992) Deletion polymorphism in the gene for angiotensin-converting enzyme is a potent risk factor for myocardial infarction. Nature 359(6396):641–644
Raynolds MV, Bristow MR, Bush EW, Abraham WT, Lowes BD, Zisman LS, Taft CS, Perryman MB (1993) Angiotensin-converting enzyme DD genotype in patients with ischaemic or idiopathic dilated cardiomyopathy. Lancet 342:1073–1075
Nomura H, Koni I, MichishitaY Morise T, Takeda R (1994) Angiotensin-converting enzyme gene polymorphism in haemodialysis patients. Lancet 343(8895):482–483
Doi Y, Yoshizumi H, Yoshinari M, Iino K, Yamamoto M, Ichikawa K, Iwase M, Fujishima M (1996) Association between a polymorphism in the angiotensin-converting-enzyme gene and microvascular complications in Japanese patients with NIDDM. Diabetologia 39(1):97–102
Kario K, Kanai N, Nishiuma S, Fujii T, Saito K, Matsuo T, Matsuo M, Shimada K (1997) Hypertensive nephropathy and the gene for angiotensin-converting enzyme. Arterioscler Thromb Vasc Biol 17(2):252–256
Lee EJ (1994) Population genetics of the angiotensin-converting enzyme in Chinese. Br J Clin Pharmacol 37(2):212–214
Evans AE, Poirier O, Kee F, Lecerf L, McCrum E, Falconer T, Crane J, O’Rourke DF, Cambien F (1994) Polymorphisms of the angiotensin-converting-enzyme gene in subjects who die from coronary heart disease. Q J Med 87(4):211–214
Pujia A, Gnasso A, Irace C, Dominijanni A, Zingone A, Perrotti N, Colonna A, Mattioli PL (1994) Association between ACE-D/D polymorphism and hypertension in type II diabetic subjects. J Hum Hypertens 8(9):687–691
Ruiz J, Blanche H, Cohen N, Velho G, Cambien F, Cohen D, Passa P, Froguel P (1994) Insertion/deletion polymorphism of the angiotensin-converting enzyme gene is strongly associated with coronary artery disease in non-insulin-dependent diabetes mellitus. Proc Natl Acad Sci USA 91(9):3662–3665
Acknowledgment
The authors gratefully acknowledge Colonel Dr. Abdulla Darweesh, Chairman of Department of Pathology in BDF hospital & Major Khalid Matar, Chief of Medical Technologists for giving the opportunity to collect blood samples, for the current study, from the BDF hospital.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Al-Harbi, E.M., Farid, E.M., Gumaa, K.A. et al. Genotypes and allele frequencies of angiotensin-converting enzyme (ACE) insertion/deletion polymorphism among Bahraini population with type 2 diabetes mellitus and related diseases. Mol Cell Biochem 362, 219–223 (2012). https://doi.org/10.1007/s11010-011-1146-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-011-1146-1