Abstract
The effect of curcumin on liver injury caused by Concanavalin A (Con A) has not been carefully examined. This study was designed to evaluate the protective effect of curcumin on Con A-induced hepatitis in mice. Liver injured mice received curcumin by gavage at a dose of 200 mg/kg body weight before Con A intravenous administration. Curcumin was effective in reducing the elevated plasma levels of aminotransferases and the incidence of liver necrosis compared with Con A-injected control group. Enzyme-linked immunosorbent assay (ELISA) showed that curcumin suppressed proinflammatory cytokines such as tumor necrosis factor (TNF)-α, interferon (IFN)-γ, and interleukin (IL)-4 production in Con A-injected mice. The reduced severity of hepatitis in curcumin pretreated mice correlated with decrease in numbers of liver CD4+ T cells but not CD8+ T cells by immunohistochemical analysis. Furthermore, the expression levels of intercellular adhesion molecule-1 (ICAM-1) and the interferon-inducible chemokine CXCL10 in hepatic tissue were significantly decreased by curcumin pretreatment. In conclusion, curcumin pretreatment protects against T cell-mediated hepatitis in mice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Excessive or misdirected immune stimulation, including T cell activation, could potentially lead to the development of liver diseases. The most common causes of life-threatening T cell-mediated liver damage in humans are infections with hepatitis B or C viruses, and autoimmune hepatitis. These hepatopathological events depend on activation of CD4+ T lymphocytes and in some case CD8+ T lymphocytes [1, 2]. Although inflammatory liver disease is a serious health problem worldwide, it can be managed pharmacologically in only a few cases.
Several well described murine models of hepatocyte injury have been established to study on the mechanisms of immune-mediated liver disease. Concanavalin A (Con A)-induced hepatitis is a well characterized model of T cell-mediated hepatitis mimicking many aspects of human T cell-mediated liver disease, including autoimmune hepatitis [2–4]. Activated CD4+ T cells, natural killer T (NKT) cells, Kupffer cells together with eosinophils infiltrate into the liver parenchyma and induce to secrete proinflammatory cytokines such as tumor necrosis factor (TNF)-α, interferon (IFN)-γ, interleukin (IL)-6, and IL-4 [3–5]. Of the cytokines involved, IFN-γ, IL-4, and TNF-α plays a critical role in direct induction of liver cytotoxicity [6–8]. It is also approved that some cytokines are tightly correlated with expression of intrahepatic adhesion as well as cytotoxic [2, 9].
Immune cells, notably CD4+ T lymphocytes, transmigration through the endothelial barrier into the inflamed tissue are prerequisite for the action of effecter cells [8, 9]. This is the adhesion cascade and distinct families of adhesion molecules and chemotactic cytokines control [9]. However, the endogenous mediators that promote the recruitment of T cells to the liver during T cell-mediated liver diseases have been poorly characterized [10].
Previous studies show that the intercellular adhesion molecule-1 (ICAM-1; CD54) and their integrin ligands contribute to hepatic CD4+ T cell accumulation induced by Con A [9, 11]. Moreover, some researchers have previously demonstrated by using flow-based adhesion assays that the interferon-inducible chemokine CXCL10 is an important in not only adhesion also in transmigration of effectors T lymphocytes through hepatic endothelium under physiological conditions of blood flow [12]. So, inhibition of lymphocyte recruitment to the liver may well represent therapeutic targets for T cell-mediated hepatitis.
Curcumin (deferuloymethane), a yellow coloring ingredient of the spice turmeric obtained from the rhizome of Curcuma longa Linn (Zingiberacea), a perennial herb distributed mainly throughout tropical and subtropical regions of the world. It has been shown to regulate numerous transcription factors, cytokines, adhesion molecules, and enzymes that have been linked to inflammation. In recent years, extensive in vitro and in vivo studies suggested curcumin has anticancer, antiviral, antiarthritic, anti-amyloid, antioxidant, and anti-inflammatory properties [13, 14]. It has been used in indigenous herbal medicine for the treatment of inflammatory and liver disorders [14, 15]. Moreover, curcumin also has immunomodulatory effects [13, 14, 16].
However, the effect of curcumin on liver injury caused by Con A has not been carefully examined. In this study, we examined the protective effect of curcumin on Con A-induced hepatitis in mice and further explored the underlying mechanisms.
Materials and methods
Mice and reagents
Pathogen-free male BALB/c mice weighing between 20 and 25 g (6 weeks old) were obtained from animal center of Fudan University (Shanghai, China). The animals were kept in an environmentally controlled room (23 ± 2°C, 55 ± 10% humidity) with a 12-h light and dark cycle and allowed free access to food and water. Fifteen to twenty mice per group were used for the present study. The study was performed in accordance with the Guiding Principles for the Care and Use of Laboratory Animals approved by the Fudan University Animal Care Committee.
Con A and curcumin were purchased from Sigma-Aldrich. The monoclonal antibodies (mAb) used in this study include anti-mouse CD4, anti-mouse CD8a (BD Pharmingen™), anti-mouse CXCL10/IP-10 (R&D Systems Inc., Minneapolis, MN), and anti-mouse ICAM-1 (Wuhan Boster Biological Technology, Ltd).
Animals treatment
Con A was dissolved in pyrogen-free saline at a concentration of 25 mg/ml and injected intravenously at a dose of 20 mg/kg body weight to induce hepatitis as described [17]. Curcumin was also dissolved in pyrogen-free phosphate buffered saline (PBS) and given by gavage at a dose of 200 mg/kg body weight. This dosage was proved effective by previous studies in mice [14].
Mice were randomly divided into four groups. Group A was the vehicle control, in which mice were not administrated Con A or curcumin, but they were given saline and PBS equally. Group B mice were orally given curcumin by gavage, without Con A treatment. Group C mice were injected tail vein with Con A, without curcumin treatment. Group D was a pretreatment group, in which mice were orally given with curcumin 40 min before challenge with Con A. The control animals in Group A, B, C were similarly handled, including oral administration of the same volume of saline and tail vein injection with the same volume of PBS.
Assay for serum transaminase activity
Blood from individual mice were obtained at 2, 8, and 24 h after Con A injection. All serum samples were collected and stored at −20°C until use. Liver enzymes including aspartate transaminase (AST) and alanine transaminase (ALT) activities in the serum were determined by spectrophotometric method with aid of automatic biochemical analyzer (Hitachi Auto Analyzer 7170, Japan).
Analysis of liver histopathology
After excision, the livers were fixed in formaldehyde. Paraffin sections were stained with hematoxylin and eosin (H&E) and examined under light microscopy. The slides were read by two of the investigators in a blinded manner.
Cytokine measurement in liver by ELISA
Cytokine (TNF-α, IFN-γ, and IL-4) levels in liver tissue were determined by enzyme-linked immunosorbent assay (ELISA) as described elsewhere [18]. For this, liver tissue was homogenized in 10 volumes of homogenization buffer containing protease mixture inhibitor (Sigma-Aldrich). After incubating for 2 h at 4°C, the homogenate was centrifuged at 12,500×g for 10 min. The supernatant was removed and centrifuged again to obtain a clear lysate. Total protein concentration of each sample using a BCA protein colorimetric assay kit (Pierce Biotechnology, USA), and samples were dispensed for ELISA kit according to the manufacturer’s instructions (R&D Systems, Inc., Minneapolis, MN). Concentration was calculated as pictograms of cytokine/milligram of total protein.
Immunohistochemistry
The liver tissue sections were dewaxed in xylene for 10 min and rehydrated in a series of graded ethanol and Tris-buffered saline (TBS) solutions. Endogenous peroxidase was blocked for 20 min (1% hydrogen peroxide in methanol), and antigen retrieval was performed by microwave heating for 15 min at 750 W in citrate buffer (pH 6.0). Sections were allowed to cool in the same buffer, washed in TBS, and blocked for 15 min at room temperature in TBS containing 10% preimmune serum from the species in which the secondary antibody was raised. CD4+ and CD8+ T cells expression was also determined in fresh frozen liver tissue. Cryostat sections were cut 5-μm thick and fixed briefly in polyformaldehyde for 10 min. Sections were blocked as above and then incubated with primary antibody. Primary antibodies were incubated on the sections overnight at 4°C at the following concentrations: rat polyclonal anti-mouse CD4 antibody, 1:25; rat polyclonal anti-mouse CD8 antibody, 1:25 dilution; rabbit anti-mouse ICAM-1, 1:100; and goat anti-mouse CXCL10 antibodies, 1:50 dilution. All antibodies were diluted in TBS–2% bovine serum albumin. Negative-control antibodies consisted of species-matched and where appropriate, immunoglobulin G (IgG) subclass-matched Ig fractions, used at the same dilution as the secondary antibodies. Incubations were performed for 30 min at room temperature. The enzyme complex was visualized by the addition of 3,3′-diaminobenzidine tetrachloride. Sections were washed with TBS between incubations.
Assessment of hepatic CD4+ and CD8+ T cells infiltration
The number of CD4+ and CD8+ positive T cells per liver was reviewed by three of the authors who were unaware of the status of animal being examined. Lymphocytes were distinguished from other leukocytes by the morphology of their nucleus. Ten microscopic fields (×400) were taken at random from each sample, and all the lymphocytes included in the field were analyzed. The mean value in the four mice was then calculated [11, 18].
Real-time quantitative PCR for ICAM-1 and CXCL10
Liver samples were collected 24 h after Con A administration. Total RNA was extracted from frozen liver tissues using TRIzol reagent according to the protocol provided by the manufacturer. For reverse transcription, we used RT-PCR kit (Perfect Real Time, SYBR® PrimeScriP™ TaKaRa). Oligonucleotide pairs for GAPDH, ICAM-1, and CXCL10 were used. The primers were as follows: ICAM-1(forward, F) 5′-CCA TCA CCG TGT ATT CGT TTCC-3′ and (reverse, R) 5′-CTGGCGGCTCAGTATCTCCTC-3′; CXCL10, (F) 5′-TGA AAT CAT CCC TGC GAG CCT AT-3′ and (R) 5′-TCC A GTT AAG GAG CCC TTT TAG ACC-3′; and GAPDH, (F) 5′-GCC TTC CGT GTT CCT ACC-3′ and (R) 5′-AGA GTG GGA GTT GCT GTT G-3′. Total RNA 1 μg was treated with DNase I to eliminate genomic DNA contamination, followed by synthesis of the first-strand using reverse transcription system. Reverse transcription was carried out as follows: 42°C for 60 min, 70°C for 10 min and first-strand cDNA was stored at −20°C. Real-time PCR was performed in a 20 μl of reaction solution containing SYBR Premix Ex Taq, primers, and cDNAs. The cycles for PCR were as follows: 95°C for 2 min, 40 cycles of 95°C for 15 s, 58°C for 20 s, and 72°C for 20 s. Melting curves were determined by heat-denaturing PCR products over a 35°C temperature gradient at 0.5°C/s from 65 to 99.5°C. GAPDH was used as an internal control. The relative amount of mRNA was determined using the △△CT technique as described previously [19]. The levels of mRNA were expressed as fold changes after normalization with GAPDH.
Statistical analysis
Data were expressed as mean ± SEM. Differences among experimental groups were determined by one-way analysis of variance (ANOVA) followed by the Tukey test when F was significant. In all comparisons, statistical significance was set at P < 0.05.
Results
Curcumin pretreatment attenuates Con A-induced hepatitis
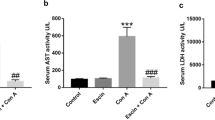
Plasma ALT and AST levels increased in response to Con A injection in mice and reached the maximal level at 8 h after Con A injection. Compared with saline or curcumin control, transaminase levels at the 8 h time-point significantly increased (P < 0.01) as shown in Table 1. Pretreatment of mice with curcumin significantly inhibited release of the transaminases ALT and AST into the plasma of Con A-treated mice (P < 0.05). Curcumin did not significantly influence transaminase in normal control mice (Table 1).
The protective effect of curcumin pretreatment was further confirmed by analysis of histological findings (Fig. 1a). As shown in the study, liver sections of control mice fixed 24 h after Con A administration and examined microscopically, exhibit inflammatory infiltration around the central veins and large areas of necrosis within the liver lobules. This histopathologic liver damage correlated with the elevated serum levels of the liver enzymes. In contrast, curcumin pretreatment dramatically reduced liver necrosis. These results show that curcumin inhibited Con A-induced liver cell death in mice.
Con A-induced liver injury is significantly inhibited by curcumin. a Photomicrographs of representative livers obtained 24 h after Con A injection with H&E staining were shown (original magnification ×200). b–d Influence of curcumin on liver cytokine levels in Con A-induced liver injury in mice. Intrahepatic TNF-α (b), INF-γ (c), and IL-4 (d) protein concentrations were quantified using ELISA at 24 h after injection of Con A to saline- or curcumin-pretreated BALB/c mice. Control animals received either saline or curcumin lone. Data are expressed as the mean ± SEM (n = 6, *P < 0.05 vs. saline/Con A)
Influence of curcumin on Con A-induced liver cytokine levels
Con A-induced hepatic injury is associated with the release of pro-inflammatory cytokines. Thus, the levels of the pro-inflammatory cytokines TNF-α, INF-γ, and IL-4 in liver tissues were assayed 24 h after Con A administration to control and curcumin pretreated mice. As shown in Fig. 1(b, c, and d), the increase intrahepatic levels of TNF-α, INF-γ, and IL-4 in response to Con A, was prevented by pretreatment with the curcumin (P < 0.05). Thus, the prevention by curcumin of Con A-induced liver injury is also associated with inhibition of proinflammatory cytokine release, for example, TNF-α, INF-γ, and IL-4.
Effect of curcumin on liver infiltrating CD4+ and CD8+ T lymphocytes
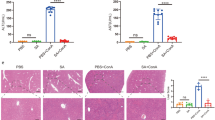
We investigated the accumulation of T lymphocytes in the liver by immunohistochemistry using anti-CD4 and CD8 stain. In normal mice, only a few infiltrating CD4+ T cells and CD8+ T cells were observed around hepatic and portal venules and sinusoids in each section. Compared with those from untreated mice, the livers of Con A-treated mice contained significantly more CD4+ T cells (Fig. 2a), but no changes in number of CD8+ T cells (Fig. 2a). Quantification of the immmunohistochemistry showed that with Con A treatment the number of CD4+ T cells per high power field (HPF) rose significantly from 3.33 ± 2.60 to 38.98 ± 24.32 (Fig. 2b). Pretreatment with curcumin significantly reduces CD4+ T lymphocyte recruitment to the liver of Con A-treated mice compared with saline control (Fig. 2b). However, liver infiltrating CD8+ T cells was not affected by curcumin (Fig. 2c). Moreover, most of the CD4+ T cells were found as large cell clusters in the periportal region.
Effect of curcumin on liver infiltrating CD4+ and CD8+ T lymphocytes. a Immunohistochemical analysis of CD4+ and CD8+ T cells infiltration in liver. Con A was administered intravenously to control mice or mice pretreated with curcumin. After 24 h, sections of livers were prepared and stained with anti-CD4+Ab or anti-CD8+Ab (original magnification, ×400). b, c Quantization of intrahepatic CD4+ and CD8+ cells 24 h after injection of Con A. CD4+ cells demonstrated a significant decrease in pretreated curcumin before Con A injection compared with control. Infiltration of CD8+ T cells was not affected by curcumin. Quantitative analysis of T cells in the liver where data are expressed as the arithmetic mean ± SEM of four mice per group. *P < 0.05 relative to saline control mice
Influence of curcumin on Con A-induced liver ICAM-1 and CXCL10 mRNA and protein
We examined intrahepatic mRNA expressions of ICAM-1 and CXCL10 at 24 h after Con A was injected. As shown in Fig. 3b and c, the intrahepatic mRNA expressions of ICAM-1 and CXCL10 in curcumin pretreated group were reduced significantly compared with saline-treated control group in Con A-induced mouse liver injury.
Influence of curcumin on Con A-induced liver ICAM-1 and CXCL10 expression. a Immunohistochemical analysis of ICAM-1 and CXCL10 in murine liver after administration of Con A for 24 h (original magnification, ×400). b, c Effect of curcumin on intrahepatic ICAM-1 mRNA and CXCL10 mRNA in Con A-induced mice. The levels of mRNA were determined at 24 h after Con A injection by real-time RT-PCR. Relative quantification of mRNA levels was calculated as the ratio between each sample and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Results are expressed as the mean ± SEM of 6 mice. The statistical differences were calculated by Student’s t test (*P < 0.05, **P < 0.01. vs. Con A-injected control group)
In normal murine liver, weak constitutive expression of ICAM-1 was observed on vascular and sinusoidal endothelial cells, and hepatocytes showed no expression of ICAM-1. ICAM-1 expression was increased on vascular and sinusoidal endothelial cells by 24 h after Con A injection (Fig. 3a). Similarly the expression of ICAM-1 and CXCL10 protein by immunocytochemical staining in liver were also reduced by curcumin (Fig. 3a).
Discussion
We examined the effect of curcumin on liver injury induced by Con A and the possible molecular mechanisms in vivo. Our results indicated that curcumin significantly inhibited the activity of serum transaminases and reduced hepatic necrosis. At the same time, curcumin significantly reduced the expression and release of several disease-relevant proinflammatory cytokines in liver induced by Con A, i.e., for TNF-α, IFN-γ, and IL-4.
Recent studies have shown that curcumin ameliorates multiple sclerosis, rheumatoid arthritis, psoriasis, and inflammatory bowel disease in human or animal models [14, 20]. In this study, we demonstrated that the curcumin effectively attenuated liver damage in a model of T cell-mediated hepatic injury. This model is characterized by an inflammatory infiltration of predominately CD4+ T cells and hepatic degeneration and necrosis 24 h after Con A administration [4, 9]. We revealed that the oral administration of curcumin significantly reduced serum levels of liver enzymes and reduced necrosis in liver compared with vehicle-treated animals.
Furthermore, TNF-α, IFN-γ, and IL-4 play crucial roles in Con A-induced hepatitis [4, 6, 7]. We noted that pretreatment of mice with a single oral dose of curcumin could significantly decrease the cytokines levels in liver 24 h after Con A-induced (Fig. 1). Previous study shown that the development of Con A-induced liver injury could be attenuated by the treatment with neutralizing anti-TNF-α or anti-IFN-γ antibodies [17]. In addition, the mice deficient of either TNFR1 or TNFR2 do not develop liver injury upon Con A stimulation [7]. Given that TNF-α and IFN-γ play major roles in Con A-induced liver injury, curcumin exerts its hepatoprotective effect through inhibition of TNF-α and IFN-γ production in this model (Fig. 1b, c). This is consistent with previous reports where curcumin treatment has profound effects on modulation of TNF-induced signaling and has been consistently shown to inhibit the expression of TNF-α and IFN-γ [20, 21]. Moreover, we also observed a significantly reduction in the IL-4 level in liver with treatment by curcumin (Fig. 1d).
During the development of inflammatory liver diseases, lymphocyte recruitment increases markedly and the intrahepatic localization of these infiltrating lymphocytes will determine the nature and severity of disease [9, 22]. Moreover, previous studies have documented CD4+ T cells are the predominant T lymphocytes recruited to the liver following Con A injection [3, 10, 17, 23]. Evidence that recruited CD4+ T cells mediate Con A-induced hepatitis in mice stems from a series of studies in which athymic nude or SCID mice were found to be resistant to Con A-induced liver damage [4, 24]. Furthermore, mice treated with anti-CD4 monoclonal antibodies (mAb) or with FK506, a T cell-specific immunosuppressant, did not develop hepatitis following Con A administration [4, 24].
In this study, we found the reduced severity of hepatitis in curcumin pretreated mice correlated with decreases in numbers of liver CD4+ T cells but not CD8+ T cells (Fig. 2). After Con A administration, the number of CD4+ T cells infiltration in liver showed increased (Fig. 2), which could contribute to the reduced cytotoxicity of CD4+ T cells from the curcumin-treated group compared with the saline-treated control group. Previous reports that curcumin has been shown to attenuate or prevent trinitrobenzene sulfonic acid-induced colitis in mice by suppressing CD4+ T cell infiltration [25]. These findings suggest that curcumin protects against Con A-induced hepatitis, at least in part, by suppressing CD4+ T cells infiltration in liver.
To further elucidate the possible causes to decrease the number of liver infiltrating CD4+ T cells, the effect of curcumin on intrahepatic ICAM-1 and CXCL10 expression in Con A-induced T cell-mediated hepatitis were examined.
Lymphocytes can enter the liver at several sites, the most important probably being the portal vessels and the sinusoids. Multiple strands of evidence implicate ICAM-1 in lymphocyte adhesion to hepatic sinusoids. It is interesting that ICAM-1 is expressed at higher levels in the liver than in other tissues [26], and the sinusoidal endothelial cells constitutively express ICAM-1 (Fig. 3a) [7, 27]. Furthermore, ICAM-1 expression on vascular and sinusoidal endothelial cells was up-regulated after 24 h Con A injection (Fig. 3a), as previously reported [7, 11]. ICAM-1 has been shown to mediate firm adhesion and to support transendothelial migration of lymphocytes in the liver [11, 28]. A previous study demonstrated that ICAM-1 deficiency significantly inhibited liver injury and CD4+ T cell recruitment in Con A-induced hepatitis [11]. Taken together, ICAM-1 expression on sinusoidal endothelial cells, as well as vascular endothelial cells, may contribute to accumulation of CD4+ T cells and subsequent liver injury in the Con A-induced hepatitis model. In this study, curcumin pretreated mice demonstrated reduced Con A-induced liver ICAM-1 mRNA and protein compared with saline control (Fig. 3). This may attribute, at least in part, to decease in numbers of CD4+ T cells recruitment in Con A-induced hepatitis.
However, pretreatment with blocking antibodies against E-selectin, P-selectin, and vascular cell adhesion molecule-1 (VCAM-1) failed to prevent Con A-induced hepatitis [7], which indicates that there are still other molecules involved in the interaction. Considering that chemokines have emerged as one of the most important regulators of leukocyte trafficking and activation [2, 22], we also assessed the effect of curcumin on intrahepatic expression of the CXCR3-associated chemokine, CXCL10 in Con A-challenged mice. It is interesting to note that we found, herein, that curcumin reduced Con A-provoked expression of CXCL10 mRNA. This decrease in CXCL10 chemokine production may partly account for the inhibitory effect of curcumin on CD4+ T cells infiltration in the liver. In fact, that notion is supported by a recent study demonstrating that CXC chemokines play a critical role in endotoxin-induced hepatic injury by inducing leukocyte extravasations [29]. Hepatic CXCL10 expression is also increased in experimental models of alcoholic liver disease. Highest expression was reported in the animals with the greatest degree of liver injury [30]. CXCL10, in common with CXCL9, is expressed in liver tissue from patients with hepatitis C infection, and relates to hepatic infiltration with CXCR3-positive T cells within the hepatic lobules [31, 32]. In contrast with the potential for CXCL10 to augment immune-mediated liver injury through the recruitment of CXCR3-positive lymphocytes, recent studies have demonstrated a hepatoprotective effect of CXCL10 [32]. But, our results showed that pretreated with curcumin reduced hepatic CXCL10 expression, which was parallel with the decreases in CD4+ T cell infiltration and liver injury. We predicted that CXCL10 and ICAM-1 expression suppressed by curcumin were likely explained in part the decreases in numbers of liver CD4+ T cells. Interestingly, there was no obvious association between CXCL10, ICAM-1 expression and CD4+ T cells infiltration in the liver. Further studies are required to elucidate true relationship between the two.
Conclusion
Curcumin exerts a hepatoprotective effect on T cell-mediated hepatitis through reduction of increased proinflammatory cytokines levels. These findings raise the possibility of using curcumin as anti-inflammatory and immunomodulatory drugs to treat inflammatory liver disease.
References
Liaw YF, Lee CS, Tsai SL et al (1995) T-cell-mediated autologous hepatocytotoxicity in patients with chronic hepatitis C virus infection. Hepatology 22(5):1368–1373
Ajuebor MN, Carey JA, Swain MG (2006) CCR5 in T cell-mediated liver diseases: what’s going on. J Immunol 177(4):2039–2045
Hong F, Jaruga B, Kim WH et al (2002) Opposing roles of STAT1 and STAT3 in T cell-mediated hepatitis: regulation by SOCS. J Clin Invest 110(10):1503–1513
Tiegs G, Hentschel J, Wendel A (1992) A T cell-dependent experimental liver injury in mice inducible by concanavalin A. J Clin Invest 90(1):196–203
Beldi G, Wu Y, Banz Y et al (2008) Natural killer T cell dysfunction in CD39-null mice protects against concanavalin A-induced hepatitis. Hepatology 48(3):841–852
Tagawa Y, Sekikawa K, Iwakura Y (1997) Suppression of concanavalin A-induced hepatitis in IFN-gamma(−/−) mice, but not in TNF-alpha(−/−) mice: role for IFN-gamma in activating apoptosis of hepatocytes. J Immunol 159(3):1418–1428
Wolf D, Hallmann R, Sass G et al (2001) TNF-alpha-induced expression of adhesion molecules in the liver is under the control of TNFR1—relevance for concanavalin A-induced hepatitis. J Immunol 166(2):1300–1307
Lafdil F, Wang H, Park O et al (2009) Myeloid STAT3 inhibits T cell-mediated hepatitis by regulating T helper 1 cytokine and interleukin-17 production. Gastroenterology 137(6):2125–2126
Shetty S, Lalor PF, Adams DH (2008) Lymphocyte recruitment to the liver: molecular insights into the pathogenesis of liver injury and hepatitis. Toxicology 254(3):136–146
Ajuebor MN, Hogaboam CM, Le T, Proudfoot AE, Swain MG (2004) CCL3/MIP-1alpha is pro-inflammatory in murine T cell-mediated hepatitis by recruiting CCR1-expressing CD4(+) T cells to the liver. Eur J Immunol 34(10):2907–2918
Kawasuji A, Hasegawa M, Horikawa M et al (2006) L-selectin and intercellular adhesion molecule-1 regulate the development of concanavalin A-induced liver injury. J Leukoc Biol 79(4):696–705
Curbishley SM, Eksteen B, Gladue RP, Lalor P, Adams DH (2005) CXCR 3 activation promotes lymphocyte transendothelial migration across human hepatic endothelium under fluid flow. Am J Pathol 167(3):887–899
Corson TW, Crews CM (2007) Molecular understanding and modern application of traditional medicines: triumphs and trials. Cell 130(5):769–774
Aggarwal BB, Harikumar KB (2009) Potential therapeutic effects of curcumin, the anti-inflammatory agent, against neurodegenerative, cardiovascular, pulmonary, metabolic, autoimmune and neoplastic diseases. Int J Biochem Cell Biol 41(1):40–59
Tang Y, Zheng S, Chen A (2009) Curcumin eliminates leptin’s effects on hepatic stellate cell activation via interrupting leptin signaling. Endocrinology 150(7):3011–3020
Xie L, Li XK, Funeshima-Fuji N et al (2009) Amelioration of experimental autoimmune encephalomyelitis by curcumin treatment through inhibition of IL-17 production. Int Immunopharmacol 9(5):575–581
Bozza M, Bliss JL, Maylor R et al (1999) Interleukin-11 reduces T-cell-dependent experimental liver injury in mice. Hepatology 30(6):1441–1447
Bonder CS, Norman MU, Swain MG et al (2005) Rules of recruitment for Th1 and Th2 lymphocytes in inflamed liver: a role for alpha-4 integrin and vascular adhesion protein-1. Immunity 23(2):153–163
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4):402–408
Siddiqui AM, Cui X, Wu R et al (2006) The anti-inflammatory effect of curcumin in an experimental model of sepsis is mediated by up-regulation of peroxisome proliferator-activated receptor-gamma. Crit Care Med 34(7):1874–1882
Fu Y, Zheng S, Lin J, Ryerse J, Chen A (2008) Curcumin protects the rat liver from CCl4-caused injury and fibrogenesis by attenuating oxidative stress and suppressing inflammation. Mol Pharmacol 73(2):399–409
Oo YH, Adams DH (2010) The role of chemokines in the recruitment of lymphocytes to the liver. J Autoimmun 34(1):45–54
Kaneko Y, Harada M, Kawano T et al (2000) Augmentation of Valpha14 NKT cell-mediated cytotoxicity by interleukin 4 in an autocrine mechanism resulting in the development of concanavalin A-induced hepatitis. J Exp Med 191(1):105–114
Mizuhara H, O’Neill E, Seki N et al (1994) T cell activation-associated hepatic injury: mediation by tumor necrosis factors and protection by interleukin 6. J Exp Med 179(5):1529–1537
Sugimoto K, Hanai H, Tozawa K et al (2002) Curcumin prevents and ameliorates trinitrobenzene sulfonic acid-induced colitis in mice. Gastroenterology 123(6):1912–1922
Park S, Murray D, John B, Crispe IN (2002) Biology and significance of T-cell apoptosis in the liver. Immunol Cell Biol 80(1):74–83
Scoazec JY, Feldmann G (1994) The cell adhesion molecules of hepatic sinusoidal endothelial cells. J Hepatol 20(2):296–300
Jaruga B, Hong F, Kim WH, Gao B (2004) IFN-gamma/STAT1 acts as a proinflammatory signal in T cell-mediated hepatitis via induction of multiple chemokines and adhesion molecules: a critical role of IRF-1. Am J Physiol Gastrointest Liver Physiol 287(5):G1044–G1052
Li X, Klintman D, Liu Q, Sato T, Jeppsson B, Thorlacius H (2004) Critical role of CXC chemokines in endotoxemic liver injury in mice. J Leukoc Biol 75(3):443–452
Simpson KJ, Henderson NC, Bone-Larson CL, Lukacs NW, Hogaboam CM, Kunkel SL (2003) Chemokines in the pathogenesis of liver disease: so many players with poorly defined roles. Clin Sci 104(1):47–63
Zeremski M, Petrovic LM, Chiriboga L et al (2008) Intrahepatic levels of CXCR3-associated chemokines correlate with liver inflammation and fibrosis in chronic hepatitis C. Hepatology 48(5):1440–1450
Koniaris LG, Zimmers-Koniaris T, Hsiao EC, Chavin K, Sitzmann JV, Farber JM (2001) Cytokine-responsive gene-2/IFN-inducible protein-10 expression in multiple models of liver and bile duct injury suggests a role in tissue regeneration. J Immunol 167(1):399–406
Author information
Authors and Affiliations
Corresponding author
Additional information
Chuan-tao Tu and Bing Han contributed equally to this work.
Rights and permissions
About this article
Cite this article
Tu, Ct., Han, B., Liu, Hc. et al. Curcumin protects mice against concanavalin A-induced hepatitis by inhibiting intrahepatic intercellular adhesion molecule-1 (ICAM-1) and CXCL10 expression. Mol Cell Biochem 358, 53–60 (2011). https://doi.org/10.1007/s11010-011-0920-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-011-0920-4