Abstract
The mitogen activated protein kinase (MAPK) signaling pathway regulates multiple events leading to heart failure including ventricular remodeling, contractility, hypertrophy, apoptosis, and fibrosis. The regulation of conserved intrinsic inhibitors of this pathway is poorly understood. We recently identified an up-regulation of Sprouty1 (Spry1) in a targeted approach for novel inhibitors of the MAPK signaling pathway in failing human hearts following reverse remodeling. The goal of this study was to test the hypothesis that up-regulated expression of Spry1 in cardiac myocytes would be sufficient to inhibit ERK1/2 activation and tissue remodeling. We established a murine model with up-regulated Spry1 expression in cardiac myocytes using the alpha-myosin heavy chain promoter (α-MHC). Heart weight and cardiac myocyte morphology were unchanged in adult male α-MHC–Spry1 mice compared to control mice. Ventricular function of α-MHC–Spry1 mice was unaltered at 8 weeks or 1 year of age. These findings were consistent with the lack of an effect of Spry1 on ERK1/2 activity. In summary, targeted up-regulation of Spry1 in cardiac myocytes is not sufficient to alter cell or tissue remodeling consistent with the lack of an effect on ERK1/2 activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Evidence to date suggests a role for the mitogen activated protein kinase (MAPK) signaling cascade in cardiac remodeling, hypertrophy, and heart failure [1, 2]. Unloading of the failing left ventricle with a left ventricular assist device (LVAD) leads to reverse remodeling, including partial normalization of myocardial structure and function by decreasing cardiac myocyte hypertrophy [3, 4]. Our lab and others have seen an association between reductions in cardiac myocyte hypertrophy and decreased activity of extracellular signal-regulated kinase (ERK1/2), following LVAD support [5–7]. We previously screened a compendium of gene expression datasets from end-stage heart failure patients before and after placement of an LVAD with the goal of identifying inhibitors of the MAPK signaling pathway [7]. From this screen, we identified Sprouty1 (Spry1), a conserved gene shown to inhibit MAPK signaling in model organisms and cells in culture. We further demonstrated that Spry1 was significantly increased after reverse remodeling of the failing left ventricle [7].
Spry1 is one of four mammalian Spry genes (Spry1–4) and was originally defined as an antagonist of branching morphogenesis in the lung of Drosophila [8]. Spry1−/− mice exhibit defects in kidney development due to increased ureteric branching, with no reported defects in cardiovascular development [9]. Studies overexpressing Spry1 in neonatal cardiac myocytes and the 293T, Swiss 3T3, C2C12, and Cos cell lines have reported decreased and delayed ERK1/2 activation in response to fibroblast growth factor-2 (FGF2) stimulation, while siRNA directed against Spry1 has been shown to increase ERK1/2 activity [7, 10, 11].
The primary goal of this study was to test the hypothesis that increased in vivo expression of Spry1 in cardiac myocytes would decrease ERK1/2 activity and alter myocyte morphology leading to decreased heart size. To test this hypothesis, we established a cardiac-specific Spry1 transgenic mouse model.
Materials and methods
Construct design and founder validation
The mSpry1 coding sequence (GenBank: AF176903) was PCR amplified from the pxj40-FLAG-Spry1 plasmid (provided by G. Guy) with primers incorporating 5′ SalI (5′-tatatagtcgacatggattccccaagtcagca-3′) and 3’ HindIII restriction sites (5′-tatataaagctttcatgacagtttgccctgag-3′). The sequence was ligated into the SalI/HindIII sites of the pnc26 plasmid (provided by J. Robbins) containing the murine α-MHC promoter (GenBank: U71441) and an hGH poly-A tail. The α-MHC/Spry1/hGH poly-A construct was excised with Not I and purified by electroelution. Transgenic founders were generated using standard pronuclear microinjection of C57/BL6 fertilized eggs implanted into foster females. PCR was performed using murine α-MHC (5′-gcccggcactcttagcaaacctca-3′) and Spry1 primers (5′-tctaacctctgccggccttccaca-3′) which amplified a 306 bp product establishing presence of the transgene; initial denature at 95°C for 4 min; 30×: denature at 94°C for 1 min, anneal at 65°C for 30 s, extend at 72°C for 30 s, and final extension at 72°C for 5 min. A Southern blot was performed with 5 μg of genomic DNA isolated from mouse liver tissue using the DNeasy Tissue Kit (Qiagen) and restriction digested with Bst XI. DNA fragments were separated by 1% PAGE and transferred to Hybond™-N+ nylon membrane (Amersham Biosciences) at RT overnight in 20× SSC buffer. A DNA probe was generated by PCR from 10 ng of α-MHC/Spry1/hGH poly-A construct using mSpry1 primers (described below in qRT-PCR). PAGE was used to confirm the correct product size (227 bp) and the probe was purified with the QiaQuick Gel Extraction Kit (Qiagen). The probe was radiolabeled with α-P32-dGTP and hybridized to the membrane overnight in Rapid-hyb™ buffer (Amersham Biosciences). The membrane was washed in 50°C 2× SSC/0.05% SDS and 0.1× SSC/0.1% SDS buffers, placed at −80°C overnight and exposed to radiography film.
Western blotting
Total protein was extracted from frozen whole mouse hearts pulverized with a modified NP40 RIPA extraction buffer containing PMSF (1 mM), NaF (50 mM), Na3VO4 (0.2 mM), BME (14.3 mM), and one complete Mini-Protease Inhibitor Cocktail tablet (Roche). Protein concentration was quantified with Bio-Rad Protein Assay (Bio-Rad), and protein was transferred to Immobilon™ PVDF membrane (Millipore) after fractionation by 10% SDS-PAGE as previously described [12]. Blots were probed overnight at 4°C with the following primary antibodies diluted in Tris-buffered saline/0.1% Tween-20 (v/v) containing 1% dried milk (Bio-Rad): anti-Spry1 (Invitrogen-Zymed; Cat. No. 40–1800), anti-Vinculin (Sigma-Aldrich; Cat. No. V9131), anti-Phospho-p44/42 MAP Kinase (Cell Signaling; Cat. No. 9101), anti-p44/42 MAP Kinase (Cell Signaling; Cat. No. 9102). HRP-conjugated goat anti-rabbit and anti-mouse secondary antibodies (Santa Cruz; Cat. No. 2004 and 2005) were used to detect their respective primary antibodies, and immunoreactive proteins were visualized by Super Signal® West Femto Maximum Sensitivity Substrate ECL detection (Pierce). Densitometry was performed using an AlphaImager® fluorimeter and analyzed with AlphaEase® FC Image Analysis Software (Alpha Innotech).
Heart weight
Mice were anesthetized by CO2 asphyxiation and immediately weighed according to the approved IACUC protocol at the University of Minnesota. Whole hearts were dissected from animals at the aortic root and rinsed three times in Dulbecco’s PBS. Hearts were blotted dry and then weighed.
Histology
Formalin-fixed mouse hearts were embedded in paraffin and 6 μm thick sections were deparaffinized with xylene and rehydrated in a graded ethanol series. Whole heart sections were stained with hematoxylin (Surgipath) for 2 min, dehydrated, and counterstained with eosin for 1 min (Surgipath). Individual cardiac myocytes were treated with TRITC-conjugated wheat germ agglutinin (Invitrogen) at 50 μg/ml for 90 min after being blocked for 60 min with 1% BSA in 1× PBS. Cardiac vascularity was determined by counting all vessels appearing in cross-section per high-power filed (400×) using TRITC stained sections. Trichrome staining was conducted according to manufacturer’s guidelines (Newcomer Supply).
Apoptosis
HL-1 cardiac myocyte cells [13] were cultured at 37°C and 5% CO2 on fibronectin (Sigma-Aldrich) coated 6-well plates (5 μg/ml) using Claycomb Media (JRH Biosciences; Cat. No. 51800C) supplemented with 10% fetal bovine serum (Gibco®), 0.1 mM norepinephrine (Sigma-Aldrich), 1% l-glutamine (Gibco®), and penicillin–streptomycin (Gibco®). HL-1 cells were infected with either eGFP or Spry1 adenovirus (described in [7]) at an MOI of 10.0 for 6 h at 37°C. 48 h later, 5 μg of Hoechst 33342 dye was added to each well for 5 min at 37°C before cells were collected by trypsinization and placed onto glass microscope slides for apoptotic scoring as previously described for condensed and coalesced nuclei [14].
Echocardiography
High resolution ultrasound biomicroscopy (VisualSonics 770, Toronto, ON) was performed using a 30-MHz linear transducer. After anesthesia was induced by inhalation of 3.5% isoflurane and supplemental oxygen, isoflurane concentration was diminished to 1.5% to maintain anesthesia. Mice were then placed upon a heated examination platform and the anterior chest was prepared for imaging with a depilatory cream. Two-dimensional (2-D) and M-mode images were obtained from standard imaging positions to determine left ventricular chamber dimensions and wall thicknesses [15, 16]. Shortening fraction and ejection fraction were calculated from M-mode and ECG-gated 2-D images, respectively [15, 16].
Statistical analysis
Statistical analyses were performed using a paired Student’s t-test. Data is presented as mean ± standard deviation. Significance was determined with 95% confidence.
Results
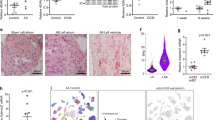
The murine Spry1 transgene was ligated in frame between the α-MHC promoter and a human growth hormone (hGH) poly-A tail (Fig. 1a). Establishing transgenic lines of founder mice was performed by Southern blot analysis (Fig. 1b). All animals were studied as heterozygous for the Spry1 transgene and followed normal Mendelian inheritance, where through five generations 55% of all male animals (n = 150) possessed the Spry1 transgene as determined by PCR-based genotyping (Fig. 1c). Spry1 protein expression was markedly increased in the hearts of transgenic animals (Fig. 1d). Spry1 overexpression was cardiac-specific and did not affect gene expression of the other Spry1 isoforms, Spry2–4 (data not shown). This initial characterization was also performed on a second founding line of transgenic mice demonstrating lesser Spry1 overexpression not discussed herein.
Characterization of the Sprouty1 transgenic mouse model. a The mouse construct design for cardiac-specific Spry1 overexpression as driven by the α-MHC promoter. b Southern blot analysis using a transgene-specific probe (solid bar in part (a)) confirmed control (CL) from transgenic (TG) animals. c PCR-based genotyping using transgene-specific primers (arrows in Fig. 1a denote primer location). d Immunoblot for Sprouty1 protein showed greater expression in transgenic (TG) mice versus control (CL) animals (n = 4)
To determine if baseline ERK1/2 activity was altered in the hearts of Spry1 transgenic mice, we performed immunoblotting for phosphorylated ERK1/2 and total ERK1/2 in 12-week old male mice (Fig. 2a). No significant differences in normalized ERK1/2 phosphorylation or total EKR1/2 expression were seen between groups following Western blotting and densitometry (Fig. 2b). Taken together, these findings suggest that in adult male mice, up-regulation of Spry1 in the heart is not sufficient to decrease basal ERK1/2 activity or alter total ERK1/2 expression levels.
Effects of Spry1 overexpression on ERK1/2 phosphorylation in the heart. a Representative immunoblot of phospho-ERK1/2 and total-ERK1/2 in control (CL) and transgenic (TG) animals. b Densitometric analysis revealed no significant differences in the degree of phospho-ERK1 relative to total-ERK1 or phospho-ERK2 relative to total-ERK2 in control (CL) (mean ± SD, n = 6) versus transgenic (TG) (mean ± SD, n = 5) animals
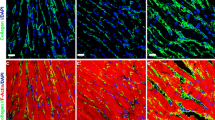
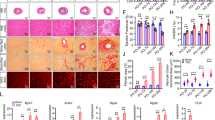
Hearts from 12-week old males demonstrated no gross morphological changes in comparison to age-matched wild-type littermate control animals (Fig. 3a). Furthermore, there was no significant difference in normalized whole heart weight between the two groups (Fig. 3b). Histological analysis of heart tissue from control and transgenic animals revealed no differences in cardiac myocyte structure (Fig. 3c) or cardiac myocyte size (Fig. 3d). Spry1 overexpression in the heart did not influence cardiac vascularity between control (CL) and transgenic (TG) animals measured as the mean number of vessels present per high-power field (CL = 8.0 ± 2.1 vs. TG = 6.0 ± 1.1, n = 5, p = NS). Additionally, cardiac fibrosis was not present in either control or transgenic animals as determined by tri-chrome staining (Fig. 3e). Finally, we did not observe any changes in cardiac myocyte morphology between control and Spry1 transgenic mice that would indicate a difference in cardiac myocyte survival (Fig. 3c). To confirm these findings, we overexpressed Spry1 in HL-1 cardiac myocytes and determined the percentage of apoptotic nuclei. Spry1 overexpression (SP) did not alter the percentage of apoptotic nuclei (CL = 1% ± 1% vs. SP = 1% ± 0%, n = 3 per group, p = NS).
Effects of Spry1 overexpression on cardiac morphometry and cardiac myocyte morphology. a Control (CL) and transgenic (TG) mouse hearts showed no differences in gross cardiac morphometry (n = 3). b There were no significant differences in normalized heart mass of control (CL) versus transgenic (TG) animals (mean ± SD, n = 6). c–e Histologic analysis revealed no deviation in control (CL) and transgenic (TG) hearts with respect to: c cardiac myocyte morphology by H&E staining (×400) (n = 5), d cardiac myocyte size or cardiac vessel number (asterisks) by WGA-TRITC staining (×400) (n = 5), or e cardiac fibrosis by tri-chrome staining (×200) (n = 5)
Structural analysis of hearts from both control and transgenic animals showed no gross differences in chamber size or wall thickness both horizontally (Fig. 4a) and longitudinally (Fig. 4b) or with regards to diastole or systole (Table 1). Functional analysis of cardiac performance by ultrasound echocardiography revealed no significant differences in either left ventricular ejection fraction or fractional shortening in 8-week old mice (Table 1). Additionally, baseline echocardiographic analysis of one-year old control (CL) versus transgenic (TG) mice demonstrated no significant changes in heart function of aged animals as determined by comparisons in ejection fraction (CL = 0.59 ± 0.00 vs. TG = 0.61 ± 0.08) and fractional shortening (CL = 0.31 ± 0.12 vs. TG = 0.29 ± 0.06).
Discussion
Spry protein family members have been classically defined by their ability to inhibit receptor tyrosine kinase (RTK) signaling [17]. Of great interest, is their potential to prevent or limit activation of the downstream RTK target ERK1/2, which is well known to play a significant role in a variety of key cellular processes in cardiac myocytes [1]. We were especially interested in Spry1 given the increased expression of this conserved ERK1/2 pathway inhibitor in failing human hearts following therapy with a LVAD that lead to reverse remodeling [7]. We hypothesized that increased expression of Spry1 in cardiac myocytes would decrease ERK1/2 activity leading to a reduction in myocyte size.
The major findings from this study are that overexpression of Spry1 in cardiac myocytes does not affect cardiovascular development, myocyte size, or ERK1/2 activity. Combined with data from the Spry1−/− mouse that did not show a cardiovascular phenotype, these data suggest that Spry1 does not play a major role under normal physiological conditions in the heart.
Previous work from our lab has shown expression of Spry1 protein in the human heart [7]. Immunohistochemical staining demonstrated the presence of Spry1 in cardiac myocytes as well as other cardiac cell types [7]. We also reported an increase in Spry1 mRNA and protein expression with an associated decrease in phosphorylated ERK1 and ERK2 in failing human hearts following unloading with a LVAD [7]. To test if up-regulation of Spry1 was causative for decreased ERK1/2 activity we overexpressed Spry1 in isolated neonatal cardiac myocytes and Cos cells and showed that acute up-regulation of Spry1 decreased the levels of phosphorylated ERK1 and ERK2 [7]. In this study, we did not see a change in phosphorylated or total ERK1 and ERK2 expression in whole hearts of Spry1 transgenic mice.
In order to elicit a change in ERK1/2 activity, our Spry1 model system might require a particular stimulus or cardiac insult. This possibility is supported by the hypothesis that Spry1 regulates developing signal transduction complexes when it becomes phosphorylated and translocates to the cell membrane [17]. As reviewed by Guy et al. [18], several key post-transcriptional mechanisms control Spry1 activity, including phosphorylation of the Y55 residue, phosphorylation within the serine-rich motif, and protein-interactions on the cysteine-rich domain. In contrast, however, it is possible that activated Spry1 was indeed present in the hearts of these animals, but its inhibition of ERK1/2 was overcome by compensatory factors acting on the Ras/MAPK signaling cascade. In particular, the hearts of these animals may have become sensitized over time to increased RTK signaling or additional signaling via G-protein coupled receptors, both of which are heavily associated with signal transduction in cardiac myocytes. It’s also possible that the effects of Spry1 can only be seen when ERK1/2 activity itself becomes increased. Work by Engelhardt et al. provide an example of this scenario, whereby without FGF2 secretion by adjacent damaged cardiac fibroblasts, there may be little increase in ERK1/2 activity for Spry1 to inhibit [19]. Finally, additional work by the Molkentin lab has shown that overexpression of the well-described dual-acting phosphatase, MKP-1, an inhibitor of ERK1/2, resulted in a decrease in cardiac myocyte hypertrophy at baseline along with decreased ventricular function [20]. However, phosphorylated ERK1/2 was not decreased in these mice at baseline, and phosphorylation of JNK1/2 and p38 were also unaffected [20].
These studies follow our initial finding of an up-regulation of the conserved ERK1/2 inhibitor, Spry1, in the failing human heart following reverse remodeling. The goal of this pre-clinical translational study was to test if Spry1 up-regulation was sufficient to inhibit ERK1/2 and decrease myocyte size. In sum, the data suggest that Spry1 up-regulation in the cardiac myocyte does not alter myocyte morphology or myocardial function.
References
Heineke J, Molkentin JD (2006) Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol 7:589–600
Wohlschlaeger J, Schmitz KJ, Schmid C, Schmid KW, Keul P, Takeda A, Weis S, Levkau B, Baba HA (2005) Reverse remodeling following insertion of left ventricular assist devices (LVAD): a review of the morphological and molecular changes. Cardiovasc Res 68:376–386
Dipla K, Mattiello JA, Jeevanandam V, Houser SR, Margulies KB (1998) Myocyte recovery after mechanical circulatory support in humans with end-stage heart failure. Circulation 97:2316–2322
Zafeiridis A, Jeevanandam V, Houser SR, Margulies KB (1998) Regression of cellular hypertrophy after left ventricular assist device support. Circulation 98:656–662
Razeghi P, Bruckner BA, Sharma S, Youker KA, Frazier OH, Taegtmeyer H (2003) Mechanical unloading of the failing human heart fails to activate the protein kinase B/Akt/glycogen synthase kinase-3beta survival pathway. Cardiology 100:17–22
Baba HA, Stypmann J, Grabellus F, Kirchhof P, Sokoll A, Schafers M, Takeda A, Wilhelm MJ, Scheld HH, Takeda N, Breithardt G, Levkau B (2003) Dynamic regulation of MEK/Erks and Akt/GSK-3beta in human end-stage heart failure after left ventricular mechanical support: myocardial mechanotransduction-sensitivity as a possible molecular mechanism. Cardiovasc Res 59:390–399
Huebert RC, Li Q, Adhikari N, Charles NJ, Han X, Ezzat MK, Grindle S, Park S, Ormaza S, Fermin D, Miller LW, Hall JL (2004) Identification and regulation of Sprouty1, a negative inhibitor of the ERK cascade, in the human heart. Physiol Genomics 18:284–289
Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA (1998) Sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell 92:253–263
Basson MA, Akbulut S, Watson-Johnson J, Simon R, Carroll TJ, Shakya R, Gross I, Martin GR, Lufkin T, McMahon AP, Wilson PD, Costantini FD, Mason IJ, Licht JD (2005) Sprouty1 is a critical regulator of GDNF/RET-mediated kidney induction. Dev Cell 8:229–239
Hanafusa H, Torii S, Yasunaga T, Nishida E (2002) Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat Cell Biol 4:850–858
Ozaki K, Miyazaki S, Tanimura S, Kohno M (2005) Efficient suppression of FGF-2-induced ERK activation by the cooperative interaction among mammalian Sprouty isoforms. J Cell Sci 118:5861–5871
Basi DL, Adhikari N, Mariash A, Li Q, Kao E, Mullegama SV, Hall JL (2007) Femoral artery neointimal hyperplasia is reduced after wire injury in Ref-1+/− mice. Am J Physiol Heart Circ Physiol 292:H516–H521
Claycomb WC, Lanson NA Jr, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, Izzo NJ Jr (1998) HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci USA 95:2979–2984
Hall JL, Matter CM, Wang X, Gibbons GH (2000) Hyperglycemia inhibits vascular smooth muscle cell apoptosis through a protein kinase C-dependent pathway. Circ Res 87:574–580
Collins KA, Korcarz CE, Lang RM (2003) Use of echocardiography for the phenotypic assessment of genetically altered mice. Physiol Genomics 13:227–239
Yang XP, Liu YH, Rhaleb NE, Kurihara N, Kim HE, Carretero OA (1999) Echocardiographic assessment of cardiac function in conscious and anesthetized mice. Am J Physiol 277:H1967–H1974
Kim HJ, Bar-Sagi D (2004) Modulation of signalling by Sprouty: a developing story. Nat Rev Mol Cell Biol 5:441–450
Guy GR, Jackson RA, Yusoff P, Chow SY (2009) Sprouty proteins: modified modulators, matchmakers or missing links? J Endocrinol 203:191–202
Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S (2008) MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456:980–984
Bueno OF, De Windt LJ, Lim HW, Tymitz KM, Witt SA, Kimball TR, Molkentin JD (2001) The dual-specificity phosphatase MKP-1 limits the cardiac hypertrophic response in vitro and in vivo. Circ Res 88:88–96
Acknowledgments
This work was supported by a Grant-in-Aid from the American Heart Association (JH, 06555827). We thank Dr. Lorrie Kirshenbaum at the University of Manitoba for his helpful discussions with this project, Dr. Graeme Guy at the National University of Singapore for providing the murine Sprouty1 plasmid, and Dr. William Claycomb of Louisiana State University for providing us with the HL-1 cardiac myocyte cell line.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Charles, N.J., Huebert, R.C., Lee, S. et al. Targeted Sprouty1 overexpression in cardiac myocytes does not alter myocardial remodeling or function. Mol Cell Biochem 342, 57–62 (2010). https://doi.org/10.1007/s11010-010-0468-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-010-0468-8