Abstract
Heat shock cognate 70 (HSC70) is an important evolutionary conserved protein that plays a major role in maintaining the homeostasis and immunity of many organisms. In this study, a HSC70 from Channa striatus was identified from its cDNA library and characterized using bioinformatics and molecular biology tools. CsHSC70 cDNA was 1953 base pair (bp) in length along with an open reading frame which encoded a polypeptide of 650 amino acid residues. Tissue distribution results showed that CsHSC70 was considerably expressed in gill, to a lesser extent in head kidney, blood, spleen and liver and at low level in other tissues. Using C. striatus gill as cell model, effects of fungal, bacterial and poly I:C stimulant on the mRNA levels of CsHSC70 was examined. We also described the antimicrobial features of two peptides namely CsHSC70 A1and CsHSC70 A2 derived from the N-terminal of CsHSC70 protein. CsHSC70 A1 peptide (40 µg/ml) exhibited potent bactericidal activity against Micrococcus luteus cells. Flow cytometric analysis revealed that the M. luteus cells stained with propidium iodide, upon treated with CsHSC70 A1 at the concentration of 40 µM/ml showed 38% survival compared to its control (99.6%). It seems that CsHSC70 A1 peptide shows antimicrobial activity against M. luteus through membrane disruption. Additionally, scanning electron microscope (SEM) observation confirmed that CsHSC70 A1 peptide treatment completely damaged and destructed the M. luteus cells. Taken together, these findings suggest that CsHSC70 A1 peptide could be a safe and potential therapeutic molecule substitute to antibiotics in various clinical fields.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Heat shock proteins (HSPs) are an evolutionarily conserved multi-gene encoded family of molecular chaperon proteins and found in many living organisms from bacteria to mammals. They play a major role in maintenance of the cellular protein homeostasis by protecting cells from a wide variety of environmental and physiological stresses, including thermal shock, oxygen radicals, heavy metals, nutrient deprivation, microbial infections, ischemia, heavy metal ions, ethanol, nicotine and surgical stress (Roberts et al. 2010; Yamashita et al. 2010). On the basis of sequence conservation and molecular size, HSPs are grouped into HSP100, HSP90, HSP70, HSP60 and low molecular weight HSP families (Roberts et al. 2010).
Among the family of heat shock proteins, the HSP70 is known to be most abundant and conserved. HSP70s are highly induced in response to a wide variety of physiological and environmental stress. However, even without stress, eukaryotic cells contain high levels of structurally related heat shock cognate proteins (HSC) encoded by constitutively active HSC70 (Liu et al. 2012). In the 70 kDa heat shock protein family, HSC70 is in the constitutively expressed form, which is actively expressed under non-stressed cells and remains unchanged or only slightly induced upon stressful stimuli, while HSP70 is highly induced during stress. HSC70 shares some of the structural and functional similarity with HSP70 and as molecular chaperon (Erbse et al. 2004; Liu et al. 2012). They play a crucial role in protein folding/unfolding, assembly/disassembly, degradation and translocation, and cellular protection against various stresses. In addition, HSC70 have also been implicated in defense response to microbial infections. HSC70 contain three domains such as an ATPase domain, a substrate-binding domain and a lid domain (Yan et al. 2010).
Antimicrobial peptides (AMPs) are major innate components of the immune system of invertebrates, vertebrates and plants against pathogens invasion. The new drugs have constantly been under investigation because of the increased development of new antibiotic-resistant bacteria (Hancock and Patrzykat 2002; Jiang et al. 2004; He et al. 2007). In recent years, naturally or designed AMPs are recognized as promising drug candidates due to their wide antimicrobial spectrum, modularity, low toxicity and decreased resistance development by target cell. All AMPs are generally small in size, membrane-active hydrophobic and positively charged molecules. The net positive charges increase their relative binding nature to microbial membrane which retains overall net negative charges. More investigation is focused on AMPs due to their high specificity to prokaryotic cells and low toxicity to eukaryotic cells (Sims et al. 1974; Nguyen et al. 2011; Sarika et al. 2012).
HSC70 and their isoforms have been cloned and characterized from various fish species and their different functions investigated against various stresses (Wang et al. 2015). However, to our knowledge no heat shock cognate proteins possess antimicrobial activity against pathogenic microorganisms. Recently, Taniguchi et al. (2013) derived three antimicrobial peptides from rice HSP70 that was found to have antimicrobial and membrane disruption activity. In this study, we have reported a complete molecular characterization of HSC70 which we identified from the cDNA library of C. striatus (Cs). The nucleotide and protein sequence of Cs HSC70 were analyzed using various biological computational tools. The temporal and spatial expression pattern of CsHSC70 in response to fungal and bacterial infection was reported. We also determined the antimicrobial features of CsHSC70 by synthesizing two peptides namely HSC70 A1 and HSC70 A2 from its amino terminal. For the first time, we report a CsHSC70 derived cationic antimicrobial peptide HSC70 A1 that exhibited potent antimicrobial activity without hemolytic activity.
Materials and Methods
CsHSC70 Identification
Full length CsHSC70 was identified from a cDNA library of C. striatus established using GS FLX™ technology. In short, cDNA library of C. striatus was developed using total RNA (Life Technologies, India), which was obtained from C. striatus. The total RNA was further purified into mRNA (Miltenyi Biotech, Germany) and then cDNA (Invitrogen, USA) was synthesized. The cDNA library was developed using Invitrogen (USA) kit and sequenced using GS FLX™ technology (Roche, India) as described in the company protocol. The detailed protocol of cDNA library development was reported in our earlier studies (Abirami et al. 2016; Prasanth et al. 2014; Rajesh et al. 2016). During Blast, a molecule encoded CsHSC70 was identified from the library and considered for the present study.
In Silico Analysis
CsHSC70 Sequence
A putative open reading frame (ORF) and its corresponding amino acid sequence of CsHSC70 cDNA was determined using Expasy Translate tool (Gasteiger et al. 2005). The physico-chemical properties of the CsHSC70 polypeptide including number of amino acid residues, molecular weight, iso-electric point (pI), instability index and aliphatic index was analyzed using PROTPARAM online server (Gasteiger et al. 2005). Also, the CsHSC70 was subjected to similarity search, which was performed through BLAST at NCBI Database. The analysis of secondary structural elements ofCsHSC70 was conducted in SOPMA (Geourjon and Deleage 1995). Themotifs and domains of the CsHSC70 were identified from PROSITE (de Castro et al. 2006), SMART (Letunic et al. 2015) and NCBI-CDD (Bauer et al. 2015) programs. Pairwise comparison and sequence alignments were carried out in Clustal W (ver. 2) (Thompson et al. 1994); further the alignment was edited in Bioedit (ver. 7.1.3.0) (Hall 1999). The evolutionary distance of CsHSC70 was obtained from Poisson model phylogenetic tree construction; for this analysis we used neighbor joining (NJ) algorithm that was embedded in MEGA 5.05 with 5000 bootstrap repetitions (Koichiro et al. 2013). Three dimensional structures of CsHSC70 proteins as well as the CsHSC70 A1 and CsHSC70 A2 peptides were established through multiple-threading alignments with potential templates (Yang et al. 2015)on I-TASSER server. The established models were visualized in PyMOL (ver. 1.00) and RasMol programs. Ramachandran plot analysis was used for the validation of the 3D models (Lovell et al. 2002).
CsHSC70 A1 and CsHSC70 A2 Peptides
The antimicrobial features such as positive charge, hydrophobic residues and protein binding potentiality of predicted peptides were predicted on PepDraw and Antimicrobial Peptide Calculator and Predictor (APD2) programs. To develop the helical wheel structure of the predicted peptides CsHSC70 A1 and CsHSC70 A2, Schiffer–Edmundson (http://rzlab.ucr.edu/scripts/wheel/wheel.cgi) program was applied.
Synthesis of Peptides
CsHSC70 derived analogs including HSC70 A1 and HSC70 A2 (Table 1) was synthesized by solid phase peptide synthesis method (SynPeptide Co., LTD, Shanghai, China). The peptides were further purified in reverse phase high-performance liquid chromatography (HPLC); and received the purity of 93.8 (HSC70 A1) and 97.1% (HSC70 A2). The molecular mass of the purified peptide analogs were confirmed by MALDI TOF-MS.
MBC of Peptide Analogs
We measured the minimum bactericidal concentration (MBC) of the two synthesized peptides against Gram-negative bacteria (Aeromonas hydrophila MTCC 1739, Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 25668,) and Gram positive bacteria (Bacillus mycoides MTCC 8920, Micrococcus luteus MTCC 6164 and Staphylococcus aureus ATCC 25923). The MBC of peptides towards selected bacteria were estimated using the modified method of Clinical and Laboratory Standards Institute (CLSI) guidelines. The selected bacteria were grown in optimum media and culture conditions overnight and diluted into each well in a density of 1 × 106 CFU/ml. In brief, 100 μl of selected bacteria were pipetted into microtiter plate with different concentrations (20, 40, 80 and 160 μg/ml) of CsHSC70 A1 and CsHSC70 A2. A negative (DEPC treated nuclease-free deionized water) and positive (Ampicillin) control were also maintained. Then, the titer plate was incubated at 37 °C for 4 h; and the MBC (μg) was determined by absorbing the optical density (OD) at 600 nm in a micro plate reader.
Quantification of Hemolytic Activity
The hemolytic activity of CsHSC70 A1 was measured according to the method described by Kumaresan et al. (2015). In short, red blood cells (RBCs) were collected from a volume of heparinized human blood (Ethical Clearance No. CDRI/IEC/2014/A1) by centrifugation at 2000×g for 5 min and washed thrice in PBS (pH 7.4) The cellular pellet was re-suspended in PBS and diluted about 4% (v/v). A 50 microliter of peptide solutions at various concentrations (20, 40, 80, 120 µg/ml) were further added and seeded in a 96-well plate. Along with this, 50 μl of 4% (v/v) Triton X-100 as positive control or 50 μl PBS as negative control was also incubated without agitation for one hat 37 °C. The hemolysis was measured by absorbance of the supernatant at 405 nm using a UV spectrophotometer (UV-1800, Shimadzu UV Spectrophotometer). The percentage of hemolysis was calculated as:
Scanning Electron Microscope (SEM) Observation
Micrococcus luteus cells were serially diluted at a density of 8 × 108 cells/ml in 200 µl of 10 mM PBS and the suspension was separated equally into two tubes. The CsHSC70 A1 (40 µg/ml) sample was added into one tube and incubated at 30 °C for 24 h. The other tube was used as a control. After the incubation, treated cells were centrifuged at 5000×g for 10 min at 4 °C and the cells prefixed in 2.5% glutaraldehyde solution and 0.1 M Na-cacodylate buffer (pH 7.2) overnight at 4 °C for 1 h. Cells were washed with series of ethanol (30, 50, 70, 90 and 100%) for dehydration. The cells were dried in liquid CO2 at a critical point. The samples were sputtered with gold using platinum coating at 10 mA for 2 min. The morphology changes of M. luteus cells treated with CsHSC70 A1 peptide were assessed using FEI Quanta FEG 200-High Resolution Scanning Electron Microscope.
Bacterial Cell Membrane Damage Induced by CsHSC70 A1
The membrane damage of CsHSC70 A1 peptide to M. luteus cells was determined by detection of propidium iodide (PI) influx into cells as done previously by Kumaresan et al. (Kumaresan et al. 2015). M. luteus cells were cultured to mid log phase and diluted to density of OD600 = 0.6 in PBS. The CsHSC70 A1 peptide (40 µg/ml) was treated with 200 µl of bacterial suspension and incubated at 37 °C for 20 min. Then, the cells were re-suspended in PI solution (5 μg/ml). The fluorescence signal in treated cells was determined by Fluorescence Assisted Cell Sorter Flow Cytometer (BD Biosciences) and analyzed with CELL-QUEST software.
Gene Expression Analysis
Animal Rearing
Healthy C. striatus with an average body weight ~50 g were received from Porurlake, Chennai, India and transported in plastic containers (50 l) to the aquaria at Division of Fisheries Biotechnology and Molecular Biology, Department of Biotechnology, SRM University, Kattankulathur. The transported individuals were acclimatized and maintained in the laboratory condition (temperature: 28 ± 1 °C and pH: 7.2 ± 0.2) as experimented in our earlier studies (Venkatesh et al. 2016; Rajesh et al. 2015; Prasanth et al. 2013; Abirami et al. 2013).
In Vivo Investigation on Immune Challenges
In order to determine the mRNA expression of CsHSC70 against fungal, bacterial and other synthetic stimulants, C. striatus were injected with the biological [(i) fungus Aphanomyces invadans, a primary causative agent of epizootic ulcerative syndrome (EUS) in murrels and (ii) bacteria Aeromonas hydrophila, a secondary causative agent of EUS in murrels] and synthetic stimulant [(iii) Poly I:C (γ-irradiated, Sigma-Aldrich, India), polyinosinic-polycytidylic acid sodium salt, a synthetic analog of double-stranded RNA (dsRNA)] intraperitoneally. PBS (1×) was prepared and served as control (100 µl/50 g fish).
In fungus challenge, each individual received 100 µl of live A. invadans at a concentration of 102 spores suspended in 1× PBS. The isolation of A. invadans from EUS infected fish was reported in our earlier studies (Prasanth et al. 2013; Abirami et al. 2013). In the bacterial challenge experiment, the fish received 100 µl of A. hydrophila per fish at a concentration of 5 × 106 CFU/ml suspended in 1× PBS. A. hydrophila was also isolated from EUS infected C. striatus muscle as reported by Dhanaraj et al. (2008). For viral analogue induction, poly I:C was injected (100 µl/50 g fish) to the fishes at a concentration of 150 µg/100 µl.
Tissue Sampling
To investigate the tissue expression of mRNA under normal physiological conditions, tissues including intestine, heart, liver, spleen, kidney, head kidney, brain, blood, gill, skin and muscle were collected from healthy individuals. Further, to study the defense responses of CsHSC70, tissues with highest expression was sampled after being challenged with pathogens and other synthetic immune stimulants at 3, 6, 12, 24, 48 and 72 h post-injection. The obtained tissues were stored as per the protocol for total RNA extraction. Three fishes were sampled for each tissue at each time point of each challenge.
RNA Extraction and cDNA Synthesis
In order to amplify the cDNA of CsHSC70, the total RNA was extracted from both the control and immune challenged fish using Tri Reagent™ method (Life Technologies, India). Invitrogen™ SuperScript™ VILO™ cDNA Synthesis Kit was used to synthesize CsHSC70 cDNA from the total RNA isolated from the tissues. Approximately 2.5 µg/µl of total RNA was used for cDNA synthesis along with 5× VILO ™ Reaction Mix (4 µl), 10× SuperScript™ Enzyme Mix (2 µl) and DEPC treated water. The contents were mixed properly and incubated at 25 °C for 10 min and 42 °C for 60 min in PCR machine. The reaction was terminated by increasing the temperature to 85 °C for 5 min. Lastly, the acquired cDNA was kept at −20 °C until further use.
Spatial and Temporal Expression by qRT-PCR Program
Gene expression pattern of CsHSC70 in different tissues from both healthy and challenged group was analyzed using Light Cycler 96 (Real Time PCR System, Roche Diagnostics GmbH, Germany). The primers designed for the study are CsHSC70 F1 (GGG ACC ATA TCT GGG CTT AAT G) and CsHSC70R2 (CAC CAC CCA GGT CAA AGA TAA G). The primers of the internal control β-actin are β-actin F3 (TCT TCC AGC CTT CCT TCC TTG GTA) and β-actin R4 (GAC GTC GCA CTT CAT GAT GCT GTT). The assay was conducted in a total volume of 20 µl including diluted cDNA (template), SYBR® Green Mix, primers and double distilled water. We followed our standardized thermal cycle condition (Prasanth et al. 2013). The obtained data were analyzed with the program supplied along with the Roche instrument and the gene expression was calculated following the method of Livak and Schmittgenm (Livak and Schmittgenm 2001).
Statistics
All the data obtained from the study were subjected to one-way ANOVA followed by Tukey’s Multiple Range Test (SPSS11.5).
Results
Identification and Analysis CsHSC70
CsHSC70 sequence was identified from the cDNA library and deposited in GenBank database under the accession ID, HF955035. The nucleotide and derived amino acid sequences of CsHSC70 are shown in E-supplementary data. The sequence comprises of 1953 base pairs (bp) nucleotides along with an open reading frame (ORF) of 650 amino acids. The predicted molecular weight of the sequence is 71.56 kDa and its theoretical isoelectric point is 5.2.
CsHSC70 Motif and Domain Analysis
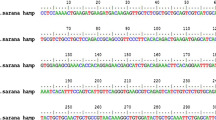
The analysis revealed that CsHSC70contained three HSP70 family signature motifs such as 9IDLGTTYS16, 97IFDLGGGTFDVSIL110 and 333IVLVGGSTRIPKIQK347. It also had three major functional domains including N-terminal ATPase domain (1–381), substrate binding domain (385–543) and C-terminal domain (537–620). The polypeptide contained acytoplasmic characteristic motif EEVD, a non-organellar stress protein motif RARFEEL and repeats of tetrapeptide (GGMP) at C terminal domain. Moreover, two bipartite nuclear localization signal motifs such as KK and RRLRT are present in the C-terminal, which is responsible for the translocation into the nucleus (Fig. 1). In addition, some high confidently predicted common motifs such as N-glycosylation site, N-myristoylation site, casein kinase II phosphorylation site, protein kinase C phosphorylation site and amidation site were also observed (E-suppl. data). In amino acid composition, the CsHSC70 polypeptide chain is rich in residues including lysine, aspartic acid, alanine, glycine, leucine, isoleucine, threonine, valine, serine and glutamine, as predicted by DNAssist 2.2.
Multiple sequence alignment of CsHSC70 with its orthologs: Siniperca chuatsi HSC70-2 (AGS83420), Seriola quinqueradiata HSC70-2 (BAG82849), S. chuatsi HSC70-1 (AGS83421), S. quinqueradiata HSC70-1 (BAG82848) and Homo sapiens HSC70 (BAD96505). The CsHSC70 containing Hsp70 family signature motifs which are enclosed in rectangular boxes. The C terminal EEVD motif was shown in single arrow underlined over the sequence. The non-organellar motif RARFEEL is indicated by block spot above the residues, and the putative bipartite nuclear localization signals, KK and RRLRT, are mentioned by ‘*’ above the sequence. The repeats of the tetrapeptide (GGMP) located in the C terminal region are underlined by double arrow over the sequence. The derived antimicrobial peptides are shaded in pink color. (Color figure online)
Homology and Phylogeny
Pair wise alignment revealed that CsHSC70 shared 96% identity to mandarin fish, Siniperca chuatsi HSC70-2 and yellowtail, Seriola quinqueradiata HSC70-2 (data not shown). Multiple sequence alignment showed that cytoplasmic characteristic motif, non-organellar stress protein motif, putative bipartite nuclear localization signal motifs such as KK and RRLRT and repeats of tetrapeptidewere strongly conserved in CsHSC70 (Fig. 1). Evolutionary relationship of the CsHSC70 was performed along with the other HSC70reported from vertebrates (E-suppl. data). The phylogenetic tree diverged into two clades; fish and higher vertebrate HSC70. The fish HSC70 clade was further separated into HSC70-1 and HSC70-2 subgroups. CsHSC70 was clustered together with HSC70-2 from Siniperca chuatsi and further grouped with HSC70-2 subgroup, which confirmed that CsHSC70 belongs to HSC70-2 isoforms. The topology of the tree mainly reflected the taxonomic classification.
Structural Characterization
The minimum free energy (MFE) of the predicted mRNA structure of CsHSC70 is -603.1 kcal/mol. The predicted mRNA structure showed that the mRNA is almost paired and very few nucleotides remain unpaired. The secondary structural elements of CsHSC70 are 36.7% α-helix, 11.8% β-sheet and 51.3% random coil. Further, the tertiary structure of CsHSC70 contained three functional domains including a 44 kDa amino-terminal adenosine triphosphatase (ATPase) domain, a 18 kDa substrate binding domain and a 10 kDa carboxyl-terminal domain (Fig. 2a). Ramachandran plot analysis was established to validate the 3D model and the results showed that 82.1% of amino acids exist in favored region, 10.3% in allowed region and 7.6% in outlier region.
Bioinformatics Analysis of CsHSC70 A1 and CsHSC70 A2 Peptides
On the basis of sequence, size, charge, charge distribution, helical propensity, hydrophobicity, amphipathicity or surface features and previous reports, we identified and derived two AMPs namely CsHSC70 A1 and CsHSC70 A2 from the N terminal of ATPase domain. Both AMPs were synthesized to analyze the antimicrobial properties. Both CsHSC70 A1 and CsHSC70 A2 were found to have a linear α-helical structure (Fig. 2b). CsHSC70 A1 showed antimicrobial properties including remarkably high positive charge (+4), hydrophobic residues (27%) and protein binding potential (3.98 kcal/mol) analyzed by PepDraw and Antimicrobial Peptide Calculator and Predictor (APD2). Moreover, the analysis showed that the amino acid percentage ofCsHSC70 A1 was found to be rich in lysine and arginine. The predicted molecular weight of CsHSC70 A1 was 2340 Daltons. Similarly, CsHSC70 A2 contains 42% hydrophobic residues and its protein binding potential was 3.81 kcal/mol. However, it is interesting to note that CsHSC70 A2 does not hold any charge (Table 1). Schiffer–Edmundson helical wheel modeling was used to predict amphipathic nature of both CsHSC70 A1 and CsHSC70 A2 (Fig. 3a, b). The wheel model showed that the peptides are linear α-helical molecule bearing both hydrophobic and hydrophilic residues on the opposing sides.
Bactericidal Activity
The bactericidal activity of the two peptides was determined by measuring minimal bactericidal concentrations (MBCs) against Gram-negative bacteria and Gram positive bacteria. Among the two, CsHSC70 A1 exhibited potent growth-inhibitory activity against Gram-positive bacteria, M. luteus (40 µg/ml) than other bacteria tested. In the case of CsHSC70 A2, it showed very low activity against M. luteus only at higher concentration (>160 µg/ml), whereas no significant activity was observed against other microorganisms tested.
Therefore, considering the potent activity of CsHSC70 A1 on the bactericidal activity, it was chosen for further assays including hemolytic assay, cell damage and membrane disruption ability.
Hemolytic Activity of CsHSC70 A1
Hemolytic activity of CsHSC70 A1 at various concentrations (20, 40, 80 and 160 µg) was determined in human RBCs. The peptide analog did not show any hemolytic activity even at a concentration of 160 µg/ml. Moreover, no difference in hemolysis was observed between CsHSC70 A1 treated RBCs and PBS treated RBCs, whereas triton X-100 showed the maximum hemolytic activity (Fig. 4).
Scanning Electron Microscopic (SEM) Observation
The morphological alteration of M. luteus cells treated with CsHSC70 A1 was determined under SEM (Fig. 5). Comparing the treated and untreated M. luteus cells under SEM, the treated cells appeared to be damaged and there was a significant destructive effect noticed which was affecting the integrity of M. luteus cells. Moreover, the outer membrane was apparently affected due to the treatment of CsHSC70 A1. Also, the treated cells become deformed in morphology with partial lack of cytoplasmic membrane. The cytoplasm of the peptide treated cells tends to spill out of the cell and ultimately lead to cell death.
Membrane Disruption Ability
Membrane disruption activity of CsHSC70 A1 on M. luteus cell membrane was determined by treating the cells with the peptide. Since PI dye is impermeable to live cells; it can be used to detect the dead cells in population. The PI can bind with DNA by intercalating between the base pairs. Hence, the cells were stained with PI and the cell morphology was determined using a flow cytometer. The assay indicated that 99.6% of cells exhibited no signal when it was not treated with peptide, whereas the PI fluorescence signal was observed when the cells were treated for nearly 20 min with peptide at a concentration of 40 µM/ml. When the cells were treated with peptide for 50 min, the percentage of cells with PI was about 38% (Fig. 6). The intensity of fluorescence increased as the concentration of the peptide increased.
Pathogen Challenge and Gene Expression of CsHSC70
Using real time PCR, relative expression of CsHSC70 mRNA was observed in all the tissues using β-actin as an internal control (Fig. 7a). However, significant expression of CsHSC70 mRNA transcripts was found in gill, to a lesser extent in head kidney, blood, spleen and liver and at low level in other tissues. Therefore, gill was chosen for further study on the possible roles of CsHSC70 mRNA regulation upon fungal, bacterial, and poly I:C stimulant. In A. invadans infected tissue, CsHSC70 expression was considerably up-regulated at 72 h post stimulation (P < 0.05) (Fig. 7b), whereas in A. hydrophila, it was significantly (P < 0.05) up-regulated at 48 h post stimulation compared to their respective controls (Fig. 7b). In poly I:C stimulated case, the expression level of CsHSC70 was started to up-regulate during 24 h (Fig. 7b) and reached its peak (P < 0.05) at 48 h post-stimulation.
Relative quantification of CsHSC70 gene expression. a Results of tissue distribution analysis of CsHSC70. Data are given as a ratio to CsHSC70 mRNA expression in skin. b The time course of CsHSC70 mRNA expression in gills at 0, 3, 6, 12, 24, 48 and 72 h post-injection with A. invadans, A. hydrophila and poly I:C. (Color figure online)
Discussion
HSC70 is the most important member of the HSP70 family, and it is constitutively expressed under non-stressed cells. It remains unchanged or slightly inducible upon stimulation. HSC70 also showed some similarity with HSP70 in structural and functional properties. Nevertheless, in comparison with HSP70 and other heat shock family members, HSC70 bears numerous unique properties. It regulates plenty of cellular functions through interaction with co-chaperones and many other molecules. It also entails in both innate and adaptive immunity including antigen presentation, T cell receptor complex formation, autoimmunity and tumor immunity and also implicates in various diseases (Liu et al. 2012; Erbse et al. 2004). It may become a biomarker for diagnosis and potential therapeutic targets for design, discovery and development of novel drugs to treat various diseases.
The CsHSC70-1 cDNA sequence was 1953 bp in length along with a 1950 bp ORF which encodes a poly peptide length of 650 amino acids. The amino acid sequence was highly homologous with its counterparts of other fish species. The sequence possesses three canonical signatures and major functional domains similar to eukaryotic HSP70 family. The non-organellar stress protein motif RARFEEL and the regulatory motif EEVD of cytoplasmic characteristic are present at the C-terminal of CsHSC70 and located in the cell cytosol and cytoplasm. The EEVD motif is strongly conserved in all eukaryotic HSC70 and other members of HSP70 family. The motif is vital for association with some co-chaperones (Demand et al. 1998; Lo et al. 2004). The putative bipartite nuclear localization signals, KK and RRLRT found in theCsHSC70 are most essential for selective translocation into nucleus (Demand et al. 1998; Liu et al. 2004).
The amino acid sequence of CsHSC70 showed a high sequence identity (96%) with HSC70-2 from S. chuatsi and S. quinqueradiata. The multiple sequence alignment of CsHSC70 with orthologous from fish and other members of HSC70 family revealed that CsHSC70 was strongly conserved (92%). Evolutionary tree classified the HSC70 into two clade; lower vertebrate (fish) and higher vertebrate HSC70s. The fish HSC70 clade was further divided into HSC70-1 and HSC70-2 as subgroup. This similar divergence was supported by Wang et al. 2015 and Yabu et al. 2011. CsHSC70 reported in this study is closely related to S. chuatsi HSC70-2. Plenty of HSC70 isoform were said to be existing in an individual genome (Liu et al. 2012). However, those different isoforms were only described in fish species including rainbow trout, zebrafish, common carp, walking catfish, yellowtail and tilapia.
The predicted mRNA structure showed that the mRNA is almost paired and very few nucleotides remain unpaired. The predicted mRNA structure of CsHSC70 along with its MFE value attempts to show that the mRNA of CsHSC70 is strongly stable. Xiong and Waterman (1997) discussed that the MFE of mRNA secondary structure based on the pairing of nucleotides. Further, they (Xiong and Waterman 1997) described that if the bases are paired they have negative value and if unpaired they have positive value. In this case, CsHSC70 mRNA had −603.1 kcal/mol, so, it is implied that the CsHSC70 mRNA has a stable structure. The total secondary structural content of CsHSC70 was 36.7% α-helix, 11.8% β-sheet and 51.3% random coil. The rich presence of random coil elements makes the tertiary structure of CsHSC70 unstructured. As in human HSC70 (Smith et al. 1998; Tsukahara et al. 2000; Sullivan et al. 2000), the tertiary structure of CsHSC70 protein is also predicted to have three functional domains including a 44 kDa amino-terminal adenosine triphosphatase (ATPase) domain, a 18 kDa substrate binding domain and a 10 kDa carboxyl-terminal domain. It is well known that HSC70 is an ATP binding chaperone, thus HSC70 showed the ATPase activity; by which it can bind and hydrolyze ATP into ADP and results in a conformational change of HSC70 and further caused substrate binding (Sullivan et al. 2000). The protein folding activity of HSC70 relies on their binding ability to expose hydrophobic amino acid sequence of the protein substrates which is regulated by ATP. The regulatory motif EEVD at the C terminal is completely conserved in HSC70 and HSP70 family members of all eukaryotes and it is the most crucial factor for association with some co-chaperones (Mosser et al. 2000).
On the basis of surface features, charges and secondary structure, two novel α-helical AMPsCsHSC70 A1 and CsHSC70 A2 were derived from CsHSC70 protein. To identify the peptides, the amino acids residues present on the surface of the CsHSC70 protein amino terminal ATPase domain were examined. The surface features were chosen because they comprise both hydrophilic and hydrophobic amino acids that make amphipathic nature which is most important for AMPs. Structural or physical parameters studies on AMPs showed numerous factors including sequence, size, charge, charge distribution, helical propensity, hydrophobicity, amphipathicity and angles subtended by hydrophobic and hydrophilic faces of the helix which control the antimicrobial activity (Giangaspero et al. 2001). CsHSC70 A1 exhibited better growth-inhibitory activity against M. luteusin lower concentration, whereas CsHSC70 A2 exhibited very low activity at higher concentration. Therefore, from the analysis it is confirmed that CsHSC70 A1 was superior to CsHSC70 A2 in hindering the growth of the pathogen. A reasonable question may arise here why the CsHSC70 A2 peptide showed very low antimicrobial activity only at higher concentration even though the peptide has amphipathicity and helical propensity; it may due to the peptide having no net charge. Also, it may due to the absence of post-translational modification in the peptide.
CsHSC70 A1 peptide showed better bactericidal activity without any hemolytic activity. There was no significant difference in hemolysis between CsHSC70 A1 and PBS treated RBCs. Hence, CsHSC70 A1 is considered to be a potent and safe AMP candidate for use against M. luteus. As PI dye is impermeable to live cells; it can generally be used for identifying cells with membrane damage in a population. The PI can bind with DNA by intercalating between the base pairs (Joshi et al. 2010; Li et al. 2013; Park and Lee 2009). Flow cytometric analysis was employed to quantitatively determine the membrane damage induced by the addition of CsHSC70 A1 to M. luteus cells. Using the technique, we found that the percentages of M. luteus cells stained with PI increased in a concentration dependent manner. The activity of CsHSC70 A1 against M. luteus was mostly due to the membrane-destabilizing ability.
The SEM observation demonstrated that CsHSC70 A1 could cause significant morphological damage to the treated bacterial cells. Treated cells appeared to be damaged and a significant destructive effect occurred that affects the integrity of M. luteus cells. The outer membrane structure is apparently affected and the cells become deformed morphologically with partial lack of cytoplasmic membrane. The cytoplasm of the cells tends to spill out of the cell and lead to death. The damage to the M. luteus cell wall and cytoplasmic membrane may cause loss of structural integrity which results in loss of cell content and cell death. Nevertheless, several probable mechanisms of AMPs against various microorganisms were proposed. Buforin II inhibits E. coli growth by means of penetrating the cell membranes and binding to DNA and RNA of cells and another AMP known as magainin 2 deters bacterial growth by forming pores on the cell membrane; despite both AMPs being linear α-helical amphipathic in nature (Park et al. 1998; Matsuzaki et al. 1997). On the basis of the obtained results, we could propose that the mode of action of CsHSC70 A1 could be akin to either buforin or magainin 2 peptide reported previously.
The expression of CsHSC70 mRNA transcripts could be detected in all the tested tissues with different expression levels. The obviously abundant CsHSC70 transcripts were observed in gill which was significantly higher than that in other tissues. So, compared with other tissues, gill may be more sensitive to stressful challenges. Also, it may due to gill being the respiratory organ and having direct contact with external or aquatic environment. The highest level of transcripts in gill proposed that CsHSC70 is used as a sensitive biomarker for different classes of environmental assault. The results of our study are in agreement with the earlier reports of Zhang et al. (2011) and Poompoung et al. (2014) who observed the expression of HSC70 and HSP70 in immune tissues including gills, head kidney, spleen and blood of grass. Moreover, it is worthwhile to note that the highest levels of CsHSC70 transcripts in gills may correlate with the osmotic regulation in striped murrel.
Accordingly, expression profiles of CsHSC70 mRNA were examined in response to challenge of A. invadans, A. hydrophilaand poly I:C induction using snakehead murrel gill as a cell model. In A. invadans infected tissue, CsHSC70 expression was considerably up-regulated at 72 h post stimulation. After poly I:C treatment, the expression level of CsHSC70 was started to up-regulate during 24 h and reached its peak at 48 h post infection. The moderate and down-regulation of CsHSC70 till 48 and 72 h post-infection of viral analog and fungal challenge suggests that the expression was seriously affected by both fungus and viral analog immunostimulant. Fungus primarily activates the host innate immune molecules through binding between PAMPs and host pattern recognition molecules (PRRs). Fungal virulent factors or PAMPs are complex carbohydrates which in turn make the pathogen-host interaction more complex which could be responsible for delayed activation of host innate immune responses (Sorrell and Chen 2009).
Upon A. hydrophila challenge, CsHSC70 was started to slightly up-regulate at 6, 24 h post challenge and peaked at 48 h post challenge compared to their respective control. The expression of HSC70-2 transcripts in the gills of walking catfish slightly increased after injection with A. hydrophila at 24 h. It was likely that CsHSC70 was insensitive to the bacteria at the early hours following injection. Bacterial infection differentially induced the expression of HSC70s in liver, gills, brain and skeletal muscle of walking catfish during 48 h. The temporal expression level of HSC70s upon A. hydrophila infection varies from organ to organ. The HSC70-2 mRNA concentrations peaked at 24 h and decreased to level of the control at 48 h in liver of walking catfish, instead,the HSC70-2 mRNA level peaked at 48 h skeletal muscle (Poompoung et al. 2014). Few more studies show that bacterial infection regulates the HSC/HSP70 mRNA transcripts in S. ocellatus (Dang et al. 2010), channel catfish (Elibol-Flemming et al. 2009), Wuchang bream (Ming et al. 2009) and Pacific abalone (Cheng et al. 2007). These findings not only revealed that HSC/HSP70 is entailed in fish immune response, but also provoked us to examine the expression patterns of HSC70 in fish immune cells to understand their roles in fish immunity. Virus also induces the heat shock response in fish and shellfishes. Reports showed that the modulation of HSP90 expression at 6, 12, 24, 48, and 72 h post infection upon nodavirus-infection in grouper fish (Chen et al. 2010). Further, Huang et al. (2008) reported that the expression of HSP21 was down-regulated upon WSSV infection in shrimp. All together, the reports showed the potential involvement of HSP/HSC against viral infection, which has been proved in our case also.
In summary, we report a complete molecular characterization of CsHSC70 which we identified from the cDNA library of C. striatus. The temporal and spatial expression pattern of CsHSC70 was determined in response to fungal, bacterial and viral analog stimulation. The nucleotide and protein sequence was analyzed using various computational tools. We also determined the antimicrobial features including bactericidal, hemolytic, cellular damage and membrane disruption activities of CsHSC70 A1 and CsHSC70 A1 peptides synthesized from amino terminal of the CsHSC70 protein sequence. Further works on the determination of the molecular mode of antimicrobial action of CsHSC70 A1 and CsHSC70 A2 has to be elucidated.
References
Abirami A, Venkatesh K, Akila S, Rajesh P, Prabha N, Prasanth B, Roy A, Thirumalai MK, Gnanam AJ, Pasupuleti M, Marimuthu K, Arockiaraj J (2013) Fish lily type lectin-1 contains β-prism 2 architecture: immunological characterization. Mol Immunol 56(4):497–506
Abirami A, Venkatesh K, Akila S, Mariadhas VA, Naif AA, Arockiaraj J (2016) Coagulation profile, gene expression and bioinformatics characterization of coagulation factor X of striped murrel Channa striatus. Fish Shellfish Immunol 55:149–158
Bauer AM, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Res 43:222–226
Chen YM, Kuo CE, Wang TY, Shie PS, Wang WC, Huang SL, Tsai TJ, Chen PP, Chen JC, Chen TY (2010) Cloning of an orange-spotted grouper Epinephelus coioides heat shock protein 90AB (HSP90AB) and characterization of its expression in response to nodavirus. Fish Shellfish Immunol 28:895–904
Cheng PZ, Liu X, Zhang GF, He JG (2007) Cloning and expression analysis of a HSP70 gene from pacific abalone (Haliotis discus hannai). Fish Shellfish Immunol 22:77–87
Dang W, Hu YH, Zhang M, Sun L (2010) Identification and molecular analysis of a stress inducible Hsp70 from Sciaenopsocellatus. Fish Shellfish Immunol 29:600–607
de Castro CJ, Sigrist A, Gattiker V, Bulliard PS, Langendijk-Genevaux E, Gasteiger E, Bairoch A, Hulo N (2006) ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34:362–365
Demand J, Luders J, Hohfeld J (1998) Thecarboxy-terminal domain of Hsc70 provides binding sites for a distinct set of chaperone cofactors. Mol Cell Biol 18:2023–2028
Dhanaraj M, Haniffa MA, Ramakrishnan CM, Singh SVA (2008) Microbial flora from the epizootic ulcerative syndrome (EUS) infected murrel Channa striatus (Bloch, 1797) in Tirunelveli region. Turk J Vet Anim Sci 32:221–224
Elibol-Flemming B, Waldbieser GC, Wolters WR, Boyle CR, Hauson L (2009) Expression analysis of selected immune-relevant genes in channel catfish during Edwardsiella ictaluri infection. J Aquat Anim Health 21:23–35
Erbse A, Mayer MP, Bukau B (2004) Mechanism of substrate recognition by Hsp70 chaperones. Biochem Soc Trans 32:617–621
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In: Walker JM (ed) The proteomics protocols handbook. Humana Press, Totowa, pp 571–607
Geourjon C, Deleage G (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci 11:681–684
Giangaspero A, Sandri L, Tossi A (2001) Amphipathic alpha helical antimicrobial peptides. Eur J Biochem 268:5589–5600
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Hancock REW, Patrzykat A (2002) Clinical development of cationic antimicrobial peptides, from natural to novel antibiotics. Curr Drug Targets 2:79–83
He J, Eckert R, Pharm T, Simanian MD, Hu C, Yarbrough DK, Qi F, Anderson MH, Shi W (2007) Novel synthetic antimicrobial peptides against Streptococcus mutans. Antimicrob Agents Chemother 51:1351–1358
Huang PY, Kang ST, Chen WY, Hsu TC, Lo CF, Liu KF, Chen LL (2008) Identification of the small heat shock protein, HSP21, of shrimp Penaeus monodon and the gene expression of HSP21 is inactivated after white spot syndrome virus (WSSV) infection. Fish Shellfish Immunol 25:250–257
Jiang S, Hong Z, Guo W, Xiaoyun, G, Gengfa, L, Yongning L, Guangxia X (2004) A synthetic peptide derived from bactericidal/permeability-increasing protein neutralizes endotoxin in vitro and in vivo. Int Immunopharmacol 4:527–537
Joshi S, Bisht GS, Rawat DS, Kumar A, kumar R, Maiti S, Pasha S (2010) Interaction studies novel cell selective antimicrobial peptides with model membranes and E. coli ATCC 11775. Biochim Biophys Acta 1798:1864–1875
Koichiro T, Glen S, Daniel P, Alan F, Sudhir K (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Kumaresan V, Bhatt P, Ganesh MR, Harikrishnan R, Arasu M, Al-Dhabi NA, Pasupuleti M, Marimuthu K, Arockiaraj J (2015) A novel antimicrobial peptide derived from fish goose type lysozyme disrupts the membrane of Salmonella enterica. Mol Immunol 68:421–433
Letunic I, Doerks T, Bork P (2015) SMART: recent updates, new developments and status in 2015. Nucl Acids Res 43:257–260
Li L, Shi YH, Cheserek MJ, Su GF, Le GW (2013) Antibacterial activity and dual mechanisms of peptide analog derived from cell-penetrating peptide against Salmonella typhimurium and Streptococcus pyogenes. Appl Microbial Biotechnol 97:1711–1723
Liu J, Yang WJ, Zhu XJ, Karouna-Renier NK, Rao RK (2004) Molecular cloning and expression of two HSP70 genes in the prawn, Macrobrachiumrosenbergii. Cell Stress Chaperon 9: 313–323
Liu T, Daniels CK, Cao S (2012) Comprehensive review on the HSC70 functions, interactions with related molecules and involvement in clinical diseases and therapeutic potential. Pharmacol Ther 136:354–374
Livak KJ, Schmittgenm TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) method. Methods 25:402–408
Lo WY, Liu KF, Liao IC, Song YL (2004) Cloning and molecular characterization of heat shock cognate 70 from tiger shrimp (Penaeus monodon). Cell Stress Chaperon 9:332–343
Lovell SC, Davis IW, Arendall WB III, de Bakker PI W, Word JM, Prisant MG, Richardson JS, Richardson DC (2002) Structure validation by Calpha RAMPAGE: Ramachandran plot assessment geometry: phi, psi and C beta deviation. Proteins 50:437450
Matsuzaki K, Sugishita K, Harada M, Fujii N, Miyajima K (1997) Interactions of an antibacterial peptide, magainin 2, with outer and inner membranes of Gram negative bacteria. Biochim Biophys Acta 1327:119–130
Ming JH, Xie J, Xu P, Liu WB, Ge XP, Liu B, He Y, Cheng Y, Zhou Q, Pan L (2009) Molecular cloning and expression of two HSP70 genes in the Wuchang bream (Megalobrama amblvcenhala Yih). Fish Shellfish Immunol 28:407–418
Mosser DD, Caron AW, Bourget L, Meriin AB, Sherman MY, Morimoto RI, Massie B (2000) The chaperone function of hsp70 is required for protection against stress-induced apoptosis. Mol Cell Biol 20:7146–7159
Nguyen LT, Haney EF, Vogel HJ (2011) The expanding scope of antimicrobial peptide strutures and their modes of action. Trends Biotechnol 29:464–472
Park C, Lee DG (2009) Fungicidal effect of antimicrobial peptide arenicin-1. Biochim Biophys Acta 1788:1790–1796
Park CB, Kim HS, Kim SC (1998) Mechanism of action of the antimicrobial peptide buforin II: buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem Biophys Res Commun 244:253–257
Poompoung P, Panprommin D, Srisapoome P, Poompuang S (2014) Cloning and expression of two HSC70 genes in walking catfish Clarias macrocephalus (Günther, 1864) challenged with Aeromonas hydrophila. Aqua Res 45:1319–1331
Prasanth B, Venkatesh K, Rajesh P, Chaurasia MK, Gnanam AJ, Mukesh P, Arockiaraj J (2013) Immunological role of C4 CC chemokine-1 from snakehead murrel Channa striatus. Mol Immunol 57:292–301
Prasanth B, Mukesh CK, Rajesh P, Venkatesh K, Abirami A, Sathyamoorthi A, Gnanam AJ, Kasi M, Pasupuleti M, Ramaswamy H, Arockiaraj J (2014) Molecular cloning, characterization and gene expression of murrel CXC chemokine receptor 3a against sodium nitrite acute toxicity and microbial pathogens. Fish Shellfish Immunol 30:245–253
Rajesh P, Venkatesh K, Harikrishnan R, Arasu MV, Al-Dhabi NA, Arockiaraj J (2015) Functional roles and gene regulation of tumor necrosis factor receptor 1 in freshwater striped murrel. Mol Immunol 66:240–252
Rajesh P, Prasanth B, Venkatesh K, Mukesh P, Arockiaraj J (2016) Innate and adaptive immune molecules of striped murrel Channa striatus. Rev Aquacult. doi:10.1111/raq.12161
Roberts RJ, AgiusC, Saliba C, Bossier P, Sung YY (2010) Heat shock proteins (chaperones) in fish and shellfish and their potential role in relation to fish health: a review. J Fish Dis 33:789–801
Sarika, Iquebal MA, Rai A (2012) Biotic stress resistance in agriculture through antimicrobial peptides. Peptides 36:322–330
Sims PJ, Waggoner AS, Wang CH, Hoffman JF (1974) Studies on the mechanism by which cyanine dyes measure membrane potential in red blood cells and phosphatidylcholine vesicles. BioChemistry 13:3315–3330
Smith DF, Whitesell L, Katsanis E (1998) Molecular chaperones: biology and prospects for pharmacological intervention. Pharmacol Rev 50:493–513
Sorrell TC, Chen SC (2009) Fungal-derived immune modulating molecules. Adv Exp Med Biol 666:108–120
Sullivan CS, Cantalupo P, Pipas JM (2000) The molecular chaperone activity of simian virus 40 large T antigen is required to disrupt Rb-E2F family complexes by an ATP-dependent mechanism. Mol Cell Biol 20:6233–6243
Taniguchi M, Ikeda A, Nakamichi S, Ishiyama Y, Saitoh E, Kato T, Ochiai A, Tanaka T (2013) Antimicrobial activity and mechanism of action of a novel cationic α-helical octadecapeptide derived from heat shock protein 70 of rice. Peptides 48:147–155
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tsukahara F, Yoshioka T, Muraki T (2000) Molecular and functional characterization of HSC54, a novel variant of human heat-shock cognate protein 70. Mol Pharmacol 58:1257–1263
Venkatesh K, Gayathri R, Faizal N, Dhayanithi NB, Arasu MV, Al-Dhabi NA, Harikrishnan R, Arockiaraj J (2016) Multifunctional murrelcaspase 1, 2, 3, 8 and 9: conservation, uniqueness and their pathogen-induced expression pattern. Fish Shellfish Immunol 49:493–504
Wang P, Xua P, Zeng S, Zhou L, Zeng L, Li G (2015) Comparative analysis of sequence feature and expression of two heat shock cognate 70 genes in mandarin fish Siniperca chuatsi. Gene 560:26–36
Xiong M, Waterman MS (1997) A phase transition for the minimum free energy of secondary structures of a random RNA. Adv Appl Math 18:111–132
Yabu T, Imamura S, Mohammed MS, TouhataK, Minami T, Terayama M, Yamashita M (2011) Differential gene expression of HSC70/HSP70 in yellowtail cells in response to chaperone-mediated autophagy. FEBS J 278:673–685
Yamashita M, Yabu T, Ojima N (2010) Stress protein HSP70 in fish. Aqua Bio Sci Monogr 3:111–141
Yan F, Xia D, Hu J, Yuan H, Zou T, Zhou Q, Liang L, Qi Y, Xu H (2010) Heat shock cognate protein 70 gene is required for prevention of apoptosis induced by WSSV infection. Arch Virol 155:1077–1083
Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) I-TASSER suite: protein structure and function prediction. Nat Methods 12:7–8
Zhang A, Zhou X, Wang X, Zhou H (2011) Characterization of two heat shock proteins (Hsp70/Hsc70) from grass carp (Ctenopharyngodon idella): evidence for their differential gene expression, protein synthesis and secretion in LPS-challenged peripheral blood lymphocytes. Comp Biochem Physiol Part B 159:109–114
Acknowledgements
This research is supported by DBT’s Prestigious Ramalingaswami Re-entry Fellowship (D.O.NO.BT/HRD/35/02/2006 and BT/RLF/Re-entry/27/2011) funded by Department of Biotechnology, Ministry of Science and Technology, Government of India, New Delhi. The authors thank King Saud University for support. We would like to thank Dr. C. Gopalakrishnan, Nanotechnology Research Centre, SRM University for his support in SEM analysis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Akila Sathyamoorthi, Rajesh Palanisamy, Mariadhas Valan Arasu, Naif Abdullah Al-Dhabi, Mukesh Pasupuleti and Jesu Arockiaraj declare that they have no conflict of interest.
Research Involving Human and Animal Rights
The animals used in this study were treated with care following the ethical procedures of the University guidelines and regulations. All experimental protocols were approved by the university research committee of SRM University. The use of human blood was approved by the ethics committee at SRM University (361/IEC/2012) and CSIR-CDRI, Lucknow (Ethical Clearance No. CDRI/IEC/2014/A1).
Informed Consent
Informed consent was obtained from the donors before collecting the blood.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sathyamoorthi, A., Palanisamy, R., Arasu, M.V. et al. Fish Heat Shock Cognate 70 Derived AMPs CsHSC70 A1 and CsHSC70 A2. Int J Pept Res Ther 24, 143–155 (2018). https://doi.org/10.1007/s10989-017-9599-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-017-9599-z