Abstract
The antimicrobial peptide from a bacterial strain is isolated from soil sample of Margalla Hills of Islamabad, Pakistan. The peptide is found to significantly inhibit the growth of both Gram-positive (Staphylococcus aureus ATCC 6538 and Micrococcus luteus ATCC 10240) and Gram-negative (Escherichia coli ATCC 25922 and Salmonella typhi ATCC 14028) bacteria as compared to gramicidin as standard. The bacterium is identified as Bacillus brevis strain MH9 based on phenotype and phylogenetic analysis. The antibacterial polypeptide was produced optimally at 35 °C after 48 h of growth, precipitated by 50 % ammonium sulphate, and further purified using HPLC. The sequential steps of purification decrease the peptide contents with prominent antibacterial activity. The peptide composed of 11 amino acid was further characterized by FT-IR and NMR. Results suggested that the peptide molecule is a novel antibacterial agent that is effective against both Gram-positive and Gram-negative bacteria. This study may have important implications for new peptide antibiotic that could be a new addition to treat infections.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The bacterial infections and resistance to broad-spectrum antibiotics are progressively increasing, demanding identification and development of new bioactive compounds against these pathogens. Since the microorganisms are reservoir of various pharmacological active compounds (Smith et al. 1997; Spellberg et al. 2008) development of antibacterial drugs from natural sources form such microbes are emerging as new trends (Jacob and Zasloff 1994).
The genus Bacillus has been extensively used in microbial and pharmaceutical biotechnology for production of antibiotics and several other important secondary metabolites (Stachelhaus et al. 1995). Several strains of Bacillus reported to produce polypeptide antibiotics including bacitracin, polymyxin, gramicidin, and colistin (Berdy 2005) under various environmental stresses (Kleerebezem and Quadri 2001). These polypeptide antibiotics are usually closely related to each other (Stachelhaus et al. 1995) and differ from each other by one or a few amino acid residues (Katz and Demain 1977).
Reported antibacterial polypeptides produced by the genus Bacilli are found effective mostly against Gram-positive bacteria (Anderson et al. 2001; Muhammad et al. 2014). However, polymyxin, colistin, and circulin show actions against Gram-negative species while bacillomycin, mycobacillin, and fungistatin are active against molds and yeasts (Anderson et al. 2001). Bacillus brevis produces gramicidin-C (Egorov 1999) which is active against Gram-positive bacteria and selectively acts against only Gram-negative Neisseria species (Bourinbaiar and Coleman 1997).
Here we report the production, purification, and characterization of a new peptide antibiotic produced by a newly isolated strain (MH9) of B. brevis that is highly effective against both the Gram-positive and Gram-negative bacteria.
Materials and Methods
Isolation, Screening, and Identification of Bacterial Strains
Strains of Mesophilic B. brevis were isolated from soil sample of Margalla Hills Islamabad, Pakistan by serial dilution method. Ten bacterial isolates were purified on nutrient agar medium at 37 °C using streak plate technique (Caccavo et al. 1994) and tested against indicator strains including Escherichia coli ATCC 25922, Salmonella typhi ATCC 14028, Micrococcus luteus ATCC 10240, and Staphylococcus aureus ATCC 6538 using agar well diffusion assay (Sen et al. 1995; Awais et al. 2008). The agar well diffusion method was used to analyze the antibacterial production by the strain. Briefly the turbidity of 24 h old cultures of indicator strains was adjusted to 0.5 McFarland turbidity standard. The test strains was then applied on the surface of Muller Hinton agar plates (30 ml) using sterilized cotton swab and permitted to dry for 15 min. We used sterilized stainless steel borer for well formation (6 mm). About 100 µl of the cell-free culture supernatants was transferred into each well. The agar plates were then incubated at required temperatures and observed for the development of zones of inhibitions around each well.
Screened bacterial isolates were morphologically characterized as described by Buchanan and Gibbons (1974). For phylogenetic characterization, genomic DNA was extracted from the bacteria using Wizard genomic Kit (Promega, Madison, USA) according to the manufacture’s instruction and PCR was performed to amplify the 16 s rRNA gene using primers 5′F (5′-AAGTCGAGCGGACAGATGG-3′) and 3′R (3′-GGGGAGGGTCATTGGAACTGG-5′). PCR was carried out in a T-Personal combi PCR machine (Biometra, Germany # 2106284) with the following programs: 3 min denaturation at 95 °C, followed by 30 cycles of 1 min denaturation at 94 °C, 1 min annealing at 58 °C, 1 min extension at 72 °C, and a final extension step of 3 min at 72 °C. PCR products of the correct sizes were purified using JET quick PCR products purification spin/250 kit (GENOMED, Germany). Sequencing of PCR products was performed and analyzed in both directions using an ABI Prism 310 automated DNA sequencer using BigDye Terminator cycle sequencing kit (PE Applied and Biosystem USA). The 16S rDNA gene sequence of bacterial strain MH9 was compared with other bacterial sequences by NCBI Basic Local Alignment Search Tool (BLASTn) for their pair wise identities. Neighbor-joining method (Saitou and Nei 1987) was used for phylogenetic tree construction and analysis using MEGA 4.0 (Tamura et al. 2007).
Production of Antibiotic by Bacillus brevis MH9 at Optimum Conditions
Bacillus brevis MH9 was grown as described by Mendo et al. (2000). The medium for the production of antibiotic was composed of l-glutamic acid: 5 g/l; KH2PO4: 0.5 g/l; K2HPO4: 0.5 g/l; MgSO4: 0.2 g/l; MnSO4: 0.01 g/l; NaCl: 0.01 g/l; FeSO4: 0.01 g/l; CuSO4: 0.01 g/l; CaCl2: 0.015; and Glucose: 1 %. The pH was adjusted to 7.5. Antibiotic was produced in a 500 ml shake-flask at 35 °C and 150 rpm agitation rate. Finally, the culture was centrifuged (10,000 rpm, 10 min, 4 °C) to separate the bacterial cells and the supernatant was stored at 4 °C (Bushra et al. 2007; Muhammad et al. 2009; Muhammad and Ahmed 2015). Optimization factors for the production of antibiotic was studied at different set of parameters: incubation time (24–144 h), agitation rate (100–150 rpm), pH (4–9), temperature (25–45 °C), glutamic acid concentration as a source of nitrogen (0.25–2 %), and carbon concentration (0.25–3 %). The antibacterial activity was confirmed using agar well diffusion assay (Sen et al. 1995).
Purification of Peptide Antibiotic
Precipitation of the peptide antibiotic was carried out using ammonium sulphate (Wingfield 2001; Charles et al. 2008). The culture supernatant was treated with powdered ammonium sulphate (20, 40, 50, and 60 % saturation). After sufficient shaking, the solution was placed on ice for 1 h and then the precipitates were collected by centrifugation at 10,000 rpm for 20 min at 4 °C. The pellet was re-suspended in 15 ml of 0.05 M potassium phosphate buffer at pH 7.5. Dialysis was carried out against the same buffer for 24 h in dialyzing bag and the pellets were freeze dried. The resultant pellets were solubilized and finally purified using isocratic elution of 40 % (v/v) acetonitrile and 0.1 % (v/v) tri-fluoroacetic acid through preparative reverse phase column chromatography HPLC (C18 column; 5 µm; 4.6 mm × 150 mm; Agilent Technologies, USA) at a flow rate of 1.0 ml/min. The collected fractions were assayed for antibiotic activity against Gram-positive S. aureus and Gram-negative E. coli using agar plate well diffusion assay (Sen et al. 1995; Awais et al. 2008). Peptide antibiotic samples were lyophilized and stored at −20 °C for further analysis.
Comparative Antibacterial Activity Analysis
Antibacterial activity of purified peptide sample obtained from B. brevis MH9 was compared to gramicidin (Sigma Aldrich) taken as standard by agar well diffusion assay.
Characterization of Peptide
The total protein content of the peptide antibiotic was estimated according to the method as described by Lowry et al. (1951). In order to determine the purity and molecular weight of the peptide, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 10 % polyacrylamide and 5 % of stacking gel (Lee et al. 1987) were used. Samples were prepared by dissolving in equal proportion of sample buffer and loaded on the wells along with molecular weight standard (spectra multicolor, page ruler low range unstained protein ladder). Half of the gel was stained with Coomassie blue dye (Lee et al. 1987) while the other half was directly assayed for antimicrobial activity following the protocol of Bhunia and Johnson (1992) to identify the peptide antibiotic on SDS-PAGE.
Amino Acid Analysis
Amino acid analyzer LC3000 was used for residual analysis of the peptide and FT-IR and NMR were used for the characterization. The FT-IR spectroscopic analysis was carried out using an Alpha FT-IR (Bruker, Germany) in the 4000–500 cm−1 region. Peptide samples for NMR study were prepared by taking 10 mg in 0.5 ml D2O and spectra were analyzed in Bruker 300 MHz spectrometer equipped with 5 mm of probe-head for 1H analysis.
Results
Finding Antibacterial Activity
The active metabolites produced by ten different isolates exhibited different degree of antibacterial activities. Among these ten isolates, the strain MH9 showed most significant results (zone of inhibition ranging from 18–28 mm) against both the Gram-positive and Gram-negative test bacteria. Most notably, the MH9 strain inhibited S. aureus growth with a zone of inhibition of 28 mm (Table 1).
Identification of Bacterial Strain
Morphologically the bacterial strain MH9 was found to be Gram-positive motile and spore forming bacilli arranged in pairs. Bacterial colony on nutrient agar plate was circular, flat and whitish in color indicating Bacillus specie. According to 16 s rRNA gene based phylogenetic analysis, the isolate is B. brevis (Accession number: X60612.1).
Phylogenetic tree with the sum of branch length 0.002 was constructed by neighbor-joining method and evolutionary relationship of sequence of query strain MH9 was determined in relation to reference sequences from NCBI (Fig. 1).
Evaluation of Peptide Antibiotic
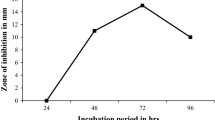
The peptide antibiotic produced by B. brevis MH9 at optimized conditions (Fig. 2) was precipitated with 50 % saturated solution of ammonium sulphate. The B. brevis MH9 indicated optimum level of peptide antibiotic production at 35 °C, 7.5 pH, 1 % glutamic acid, and 1.5 % glucose concentration after 48 h of incubation. The precipitated peptide antibacterial compound was isolated and purified from the supernatant. The concentration of peptide sample was estimated and its antibacterial activity was measured. The details of these analyses are represented in Table 2. It was observed that additional purification steps increase the purity of the peptide, but reduce the yield of the peptide. The HPLC chromatogram of active fractions of peptide samples showed retention time of 24.04 min at 254 nm (Fig. 3).
Comparative Analysis of Antibacterial Activity
The purified peptide sample showed prominent antibacterial activity with a zone of inhibition of 34 and 28 mm against S. aureus and E. coli respectively as compared to gramicidin as standard which indicated 22 and 2 mm zone of inhibition respectively against both test bacterial strains (Fig. 4).
Characterization of the Peptide Antibiotic
The amino acid components of the purified peptide antibiotic fraction from MH9 revealed that it contains 11 amino acid residues (Table 3). SDS-PAGE showed a single band at position below 1.6 kDa marker (~1.4 kDa) (Fig. 5). The FT-IR spectrum of peptide antibiotic showed absorption peaks at the regions of 794 cm−1 (C=C), 1257 cm−1 (C–O), 1700 cm−1 (N–H), 1940 cm−1 (O–N–O), 2350 cm−1 (–C=N), 2810 cm−1 (=C–H), 3430 cm−1 (–N–C–H), 3490 cm−1 (H–O–H), and in the region of 3620 cm−1 (–OH) (Fig. 6). The 1H NMR spectral signals demonstrated that the peptide is having both the aliphatic and aromatic groups: H-NMR (300 MHZ, 5 ppm): 7.25–7.37 (m, 4H, Ar–H), 5.37 (s, 2H, NH2), 10.3 (bS, 1H, OH), and 13.9 (S, 1H).
Discussion
Bacillus species are known to produce secondary metabolites that are the object of natural product chemistry studies. Their antimicrobial activities have motivated the pharmaceutical industry to search for lead structures in microbial extracts. Bacterial secondary metabolites including peptides antibiotics (Zuber and Marahiel 1997) are getting importance because of their increasing clinical usages (Schallmey et al. 2004). These polypeptides are having several advantages such as greater efficacy against broad spectrum multi-drug resistant bacterial pathogens among other uses (Augustin et al. 2009). The genus Bacillus is known for its antimicrobial peptides for decades against various microbial infections and industrial uses (Zhang and Falla 2004; Abriouel et al. 2011) and Bacillus subtilis reported to produce antimicrobial lipopeptides (Touré et al. 2004; Chen et al. 2008).
We have isolated Bacillus strains from Margalla Hills which is also found in some other geographical locations (Hill et al. 2004; Priest et al. 2004). Like the other Bacillus (Yasawong et al. 2011), our selected strain MH9 is morphologically Gram-positive, rod-shaped, aerobic, motile, and spore forming.
Most of the Bacilli are mesophilic that produce peptide antibiotics (gramicidins, tyrocidines, and bacitracins) (Egorov et al. 1987), while few of them are capable of growing at temperature above 40 °C. Such strains are B. brevis var. G-B that produces gramicidin C (Egorov 1999) and Bacillus polymyxa that manufactures gavaserin and saltavalin (Pichard et al. 1995). Gramicidins produced by B. brevis strains are heterogeneous mixture of polypeptide antibiotics (Bourinbaiar and Coleman 1997) that contains ~15 amino acid residues (Burkhart et al. 1999). Similarly, tyrocidine is the major constituent of tyrothricin, which also contains gramicidin. It is a mixture of cyclic-decapeptides obtained from B. brevis and contains 4-different amino acid sequences. Tyrocidine has been reported to be toxic to human blood and reproductive cells (Mootz and Marahiel 1997). While in our study, the isolated polypeptide antibiotic from the strain MH9 was found to contain 11 amino acid residues. Most of the antimicrobial peptides produced by Bacillus either acts against Gram-positive or Gram-negative pathogens (Anderson et al. 2001). Gramicidin, for example has been reported against Gram positive and selectively against Gram negative Neisseria species, while tyrocidine has been used only against localized Gram-positive bacterial infections (Bourinbaiar and Coleman 1997; Robertson and Maibach 1998). But, we observed that our isolated peptide from MH9 strain is equally active against both the Gram-positive and Gram-negative test bacteria. The significant antibacterial activity of MH9 sample with a maximum zone of inhibition of 28 mm was observed against E. coli as compared to gramicidin which indicated 2 mm zone of inhibition (Robertson and Maibach 1998) against the same bacterial species.
Antibiotic production by bacteria varies quantitatively and qualitatively depending on the culture conditions (Chen et al. 1996; Fang et al. 1997) and strain (Berditsch et al. 2007). Important factors in culture conditions include-the time of incubation, pH, temperature, aeration rate, and chemical components including nitrogen, and carbon concentrations (Awais et al. 2008; Webster et al. 2002). B. subtilis strain SK.DU.4 is reported to produce antimicrobial peptides under 14 % NaCl after 24 h of growth (Baindara et al. 2013). In our case, the B. brevis MH9 exhibited optimum level of peptide antibiotic production at 35 °C after 48 h of incubation. The optimum pH, glutamic acid, and glucose concentration were 7.5, 1, and 1.5 %, respectively.
The first step in antibiotic purification is separation of crude antibiotic from the microbial growth followed by precipitation of proteins by 70 % ammonium sulfate (Shimogki et al. 1991). Subsequently, HPLC can be used for the purification and collection of active fractions containing bacitracin and other antibacterial polypeptides (Sunaryanto et al. 2010). Using a strong isocratic-HPLC technique Oka and co-workers (Oka et al. 1989) had purified 22 bacitracin constituents in isocratic conditions with 25 min run time while in current study, the peptide antibiotic from B. brevis strain MH9 was precipitated by 50 % saturated solution of ammonium sulphate and further purified by reverse phase column HPLC. The purified antibiotic peptide is ~1.4 kDa which is in agreement with a previous study where gramicidin from some B. brevis strain is reported to have molecular weight less than 2 kDa (Woolley and Wallace 1994).
FT-IR spectra of gramicidin components were recorded in the region of 1000–3750 cm−1 (Zuber and Marahiel 1997) while the spectra of peptide antibiotic produced by B. brevis MH9 was recorded in the region of 794–3620 cm−1 supporting the finding of Rijs et al. (2011). These findings showed that peptide antibiotic may fall under gramicidin class. 1H NMR spectra also suggested that the peptide sample contains alkyl groups, amide linkage, and carbonyl groups and is composed of amino acids: Gly, Leu, Trp, Tyr, Lys, Ile, Ala, Phe, Arg, Asp, and Pro. We studied a new peptide antibiotic produced by B. brevis strain MH9 with molecular weight of ~1.4 kDa. It is effective against Gram-positive and Gram-negative bacteria as compared to previously reported gramicidin and tyrocidine antibiotics. The peptide contains 11 amino acids residues however the exact sequence of the polypeptide chain is yet to be determined and also the gene that is translated into this peptide.
Conclusion
With increased emergence of bacterial resistance depicts the need of antimicrobial drug development. Results of this study suggested that the peptide molecule is a novel antibacterial agent that is effective against both Gram-positive and Gram-negative bacteria. This study may have important implications for new peptide antibiotic that could be a new addition to treat infections. It will be helpful in projecting low production cost, less toxicity and promising pharmacokinetics to develop highly effective antimicrobial peptides with high biological activities.
References
Abriouel H, Franz CM, Ben Omar N, Gálvez A (2011) Diversity and applications of Bacillus bacteriocins. FEMS Microbiol Rev 35:201–232
Anderson A, O’Neil R, Surti TR (2001) Stroud, approaches to solving the rigid receptor problem by identifying a minimal set of flexible residues during ligand docking. Chem Biol 8:445–457
Augustin R, Anton-Erxleben F, Jungnickel S et al (2009) Activity of the novel peptide arminin against multi-resistant human pathogens shows the considerable potential of phylogenetically ancient organisms as drug sources. Antimicrob Agents Chemother 53:5245–5250
Awais M, Pervez A, Qayyum S, Saleem M (2008) Effects of glucose, incubation period and pH on the production of peptide antibiotics by Bacillus pumilus. Afr J Microbiol Res 2:114–119
Baindara P, Mandal SM, Chawla N, Singh PK, Pinnaka AK, Korpole S (2013) Characterization of two antimicrobial peptides produced by a halotolerant Bacillus subtilis strain SK.DU.4 isolated from a rhizosphere soil sample. AMB Express 3:2
Berditsch M, Afonin S, Ulrich AS (2007) The ability of Aneurinibacillus migulanus (Bacillus brevis) to produce the antibiotic gramicidin S is correlated with phenotype variation. Appl Environ Microbiol 73:6620–6628
Berdy J (2005) Bioactive microbial metabolites. A personal view. J Antibiot 58:1–26
Bhunia AK, Johnson MG (1992) A modified method to directly SDS-PAGE the bacteriocin of Pediococcus acidilactici. Lett Appl Microbiol 15:5–7
Bourinbaiar AS, Coleman CF (1997) The effect of gramicidin, a topical contraceptive and antimicrobial agent with anti-HIV activity, against herpes simplex viruses type 1 and 2 in vitro. Arch Virol 142:2225–2235
Buchanan JR, Gibbons NE (1974) Bergey’s manual of determinative bacteriology, 8th edn. Williams and Wilkins Company, Baltimore
Burkhart BM, Gassman RM, Langs DA, Pangborn WA, Duax WL, Pletnev V (1999) Gramicidin D conformation, dynamics and membrane ion transport. Biopolymers 51:129–144
Bushra J, Hasan F, Hameed A, Ahmed S (2007) Isolation of Bacillus subtilis MH-4 from soil and its potential of polypeptide antibiotic production. Pak J Pharm Sci 20:26–31
Caccavo F Jr, Lonergan DJ, Lovley DR, Davis M, Stolz JF, McInerney MJ (1994) Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl Environ Microbiol 60:3752–3759
Charles P, Devanathan V, An bu P, Ponnuswamy MN, Kalaichelvan PT, Hur BK (2008) Purification, characterization and crystallization of an extracellular alkaline protease from Aspergillus nidulans HA-10. J Basic Microbiol 48:347–352
Chen G, Maxwell P, Dunphy GB, Webster JM (1996) Culture conditions for Xenorhabdus and Photorhabdus symbionts of entomo pathogenic nematodes. Nematologica 42:124–127
Chen H, Wang L, Su CX, Gong GH, Wang P, Yu ZL (2008) Isolation and characterization of lipopeptide antibiotics produced by Bacillus subtilis. Lett Appl Microbiol 47:180–186
Egorov NS (1999) Vestn. Mos. Gos. Univ. Ser Biol 4:38–49
Egorov NS, Silaev AB, Katrukha GS et al (1987) Antibiotiki-polipeptidy (polypeptide anyibiotics). Mosk. Gos. Univ, Moscow, p 263
Fang A, Pierson DL, Mishra SK, Koenig DW, Demain AL (1997) Gramicidin S production by Bacillus brevis in simulated microgravity. Curr Microbiol 34:199–204
Hill KK, Ticknor LO, Okinaka RT, Asay M, Blair H, Bliss KA et al (2004) Fluorescent amplified fragment length polymorphism analysis of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis isolates. Appl Environ Microbiol 70:1068–1080
Jacob L, Zasloff M (1994) Potential therapeutic applications of magainins and other antimicrobial agents of animal origin. In: Boman HG, Marsh J, Goode JA (eds) Antimicrobial peptides, ciba foundation symposium. Willey, Chichester, p 186
Katz E, Demain A (1977) The peptide antibiotics of Bacillus: chemistry, biogenesis, and possible functions. Bacteriol Rev 41:449–474
Kleerebezem M, Quadri LE (2001) Peptide pheromone-dependent regulation of antimicrobial peptide production in gram-positive bacteria: a case of multicellular behavior. Peptides 22:1579–1596
Lee C, Levin A, Branton D (1987) Copper staining: a five-minute protein stains for sodium dodecyl sulfate-polyacrylamide gels. Anal Biochem 166:303–312
Lowry OH, Rosenbrough NJ, Farr AL, Randall RJ (1951) Protein measurement with folin phenol reagent. J Riol Chem 193:265–275
Mendo S, Henriques I, Correia A, Duarte J (2000) Genetic characterization of a new thermotolerant Bacillus licheniformis strain. Curr Microbiol 40:137–139
Mootz HD, Marahiel MA (1997) The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J Bacteriol 179:6843–6850
Muhammad SA, Ahmed S (2015) Production and characterization of a new antibacterial peptide obtained from Aeribacillus pallidus SAT4. Biotechnol Rep 8:72–80
Muhammad SA, Ahmad S, Hameed A (2009) Antibiotic production by thermophilic Bacillus species SAT-4. Pak J Pharm Sci 22:339–345
Muhammad SA, Ahmed S, Ismail T, Hameed A (2014) Taguchi’s experimental design for optimizing the production of novel thermostable polypeptide antibiotic from Geobacillus pallidus SAT4. Pak J Pharm Sci 27:11–23
Oka H, Harada K, Suzuki M, Nakazawa H, Ito Y (1989) Foam counter-current chromatography of bacitracin. I. Batch separation with nitrogen and water free of additives. J Chromatogr 482:197–205
Pichard B, Larue JP, Thouvenot D (1995) Gavaserin and saltavalin, new peptide antibiotics produced by Bacillus polymyxa. FEMS Microbiol Lett 133:215–218
Priest FG, Barker M, Baillie LWJ, Holmes EC, Maiden MCJ (2004) Population structure and evolution of the Bacillus cereus group. J Bacteriol 186:7959–7970
Rijs AM, Kabelac M, Abo-Riziq A, Hobza P, de Vries MS (2011) Isolated gramicidin peptides probed by IR spectroscopy. Chem Phys Chem 12:1816–1821
Robertson DB, Maibach HI (1998) Dermatologic pharmacology. In: Katzung BG (ed) Basic and clinical pharmacology. Appleton-Lange, Norwalk, p 1000
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schallmey M, Sing A, Ward OP (2004) Developments in the use of Bacillus specie for industrial production. Can J Microbiol 50:1–17
Sen S, Sahu NP, Mahato SB (1995) Pentacyclic triterpenoids from Mimusops elengi. Phytochem 38:205–207
Shimogki H, Takeuchi K, Nishino T, Ohdera M, Kudo T, Ohba K, Iwnma M, Irie M (1991) Purification and properties of a novel surface active agent and alkaline resistant protease from Bacillus sp. Agric Biol Chem 55:2251–2258
Smith NK, Gilmour SG, Rastall RA (1997) Stational optimization of enzymatic synthentic of derivatives of tetrabore and sucrose. Enzyme Microbial Technol 21:349–354
Spellberg BR, Guidos D, Gilbert J, Bradley HW, Boucher WM, Scheld JG, Edwards BJ (2008) The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis 46:155–164
Stachelhaus T, Schneider A, Marahiel M (1995) Rational design of peptide antibiotics by targeted replacement of bacterial and fungal domains. Science 269:69–72
Sunaryanto R, Marwoto B, Irawadi TT, Masud ZA, Hartoto L (2010) Antibiotic compound from marine actinomycetes (Streptomyces sp A11): isolation and structure elucidation. Indo J Chem 10:219–225
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Touré Y, Ongena M, Jacques P, Guiro A, Thonart P (2004) Role of lipopeptides produced by Bacillus subtilis GA1 in the reduction of grey mould disease caused by Botrytis cinerea on apple. J Appl Microbiol 96:1151–1160
Webster JM, Chen G, Hu K, Li J (2002) Bacterial metabolites. In: Gaugler R (ed) Entomo pathogenic nematology. CAB International, Wallingford, pp 99–114
Wingfield P (2001) Protein precipitation using ammonium sulfate. Curr Prot Protein Sci A.3F.1–A.3F.8
Woolley GA, Wallace BA (1994) Membrane protein structure: lessons from gramicidin., Methods in Physiology SeriesSpringer, New York, pp 314–334
Yasawong M, Areekit S, Pakpitchareon A, Santiwatanakul S, Chansiri K (2011) Characterization of thermophilic halotolerant Aeribacillus pallidus TD1 from Tao dam hot spring, Thailand. Int J Mol Sci 12:5294–5303
Zhang L, Falla TJ (2004) Cationic antimicrobial peptides—an update. Exp Opin Investig Drugs 13:97–106
Zuber PA, Marahiel MA (1997) In: Strohl W (ed) Biotechnology of antibiotics. Marcel Dekker, New York, pp 187–216
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
All authors declare that they have no conflict of interest.
Funding
There was no funding available for this research. We acknowledge the administrative support of Atta-ur-Rahman School of Applied Biosciences, National University of Sciences and Technology, Islamabad and Institute of Molecular Biology and Biotechnology, Bahauddin Zakariya University Multan, Pakistan.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Muhammad, S.A., Ali, A., Naz, A. et al. A New Broad-Spectrum Peptide Antibiotic Produced by Bacillus brevis Strain MH9 Isolated from Margalla Hills of Islamabad, Pakistan. Int J Pept Res Ther 22, 271–279 (2016). https://doi.org/10.1007/s10989-015-9508-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-015-9508-2