Abstract
Context
Landscape genetics focuses on quantifying relationships between landscape features and gene flow. Most past landscape genetic studies have addressed relationships between landscape structure and genetic differentiation in single study areas. However, failure to investigate multiple areas across a species range could produce misleading inferences about which landscape variables generally limit gene flow in a given species resulting in faulty management decisions.
Objectives
The main objectives of this paper were to identify the landscape features that facilitate or impede stone marten gene flow across its Iberian range, and to test whether gene flow is always influenced by the same set of landscape features or if they are detected only when they are limiting in a particular landscape.
Methods
We conducted an individual-based meta-replicated landscape genetic analysis using multivariate-restricted optimization with reciprocal causal modeling in three study areas within the Iberian Peninsula with strongly contrasting landscape characteristics.
Results
Variables explaining the stone marten distribution differed among areas, confirming that relationships between genetic connectivity and landscape features are variable across Iberia. We found clear patterns in which variables were included in the final optimized model related to how limiting those variables were to gene flow in each particular study area.
Conclusions
This study highlights that variability in limiting factors can have a large effect on predictions of which landscape features affect gene flow. Our results suggest that landscape models specific to the region of interest should be developed before proposing management and conservation actions, and contribute to the growing understanding of limiting factors in landscape genetics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Landscape genetics combines population genetics, landscape ecology and spatial statistics (Manel et al. 2003; Balkenhol et al. 2015). In individual-based landscape genetic analyses, the individual is used as the operational unit, avoiding potential biases associated with the delimitation of pre-defined populations (Schwartz and McKelvey 2009; Cushman et al. 2015), and often providing higher statistical power and spatial resolution (Manel et al. 2003; Landguth et al. 2010).
The predominant approach to identifying relationships between landscape features and genetic differentiation used in landscape genetics today is based on correlations between pairwise individual genetic distances (GDs) and effective distances among individuals across the landscape mosaic. The goal is to quantify the effect that landscape composition, configuration and quality have on micro-evolutionary processes and on the spatial patterns of genetic variability (Balkenhol et al. 2009, 2015). There has been controversy over the use of Mantel tests in landscape genetics (Balkenhol et al. 2009; Guillot and Rousset 2011, 2013; Graves et al. 2012, 2013), but an alternative has yet to be identified that does not also suffer drawbacks. Shirk et al. (2010) proposed a restricted multivariate optimization approach that uses Mantel r value as the objective function being optimized across combinations of resistance parameterizations. Castillo et al. (2014) showed using simulations that the Shirk et al. (2010) multivariate optimization using the reciprocal causal modeling approach proposed by Cushman et al. (2013b) identified the true drivers of genetic differentiation and produced useful resistance estimates. The approach is based on relative support (RS) of each candidate model in competition with all other candidate models at each step of a restricted multivariate optimization. Recently, Zeller et al. (2016) found that restricted multivariate optimization with reciprocal causal modeling provided better identification of factors driving gene flow than several other approaches. Furthermore, in one of the most comprehensive simulation studies of landscape genetic methods conducted to date, Shirk et al. (2017) found that Mantel-based models selection methods performed very well in identifying the correct driving variables and their relative influence. In addition, the approach we have used here seems to avoid most of the issues associated with spatial autocorrelation identified by Guillot and Rousset (2011). First, the main issue identified by Guillot and Rousset is that spatial autocorrelation inflates Type I error rates by affecting p-values. However, our approach does not use p-values, but rather optimizes the objective function of the relative magnitude of the partial Mantel r value. Second, Guillot and Rousset showed that the absolute values of the correlation coefficients were affected, but not the relative magnitudes. Therefore optimizing based on the relative magnitude of the partial Mantel r value likely avoids these issues. In addition, while there have been a number of other approaches proposed for landscape genetic analysis and optimization of resistance surfaces, such as Bedassle (Bradburd et al. 2013) and Sunder, most of them have not been well vetted with simulation analysis. It was not the purpose of this paper to evaluate the relative performance of such methods, but to apply a well known and widely applied method to a meta-replicated analysis of stone-marten landscape genetics on the Iberian Peninsula.
One of the key ideas in landscape ecology to emerge over the past two decades is the importance of landscape-level replication to determine generality and variation in pattern–process relationships (e.g., Turner and Gardner 1991; Hargrove and Pickering 1992; Wiens et al. 1993; McGarigal and Cushman 2002; Radford et al. 2005). Factors that affect species distributions (e.g., Shirk et al. 2014) and gene flow (e.g., Short Bull et al. 2011) may differ among study areas, with certain factors limiting distributions or gene flow in some landscapes while other factors are seen to be important elsewhere. Reliable inferences about relationships between landscape patterns and population processes require replicated studies at the landscape-level to identify general patterns and quantify threshold relationships where landscape features begin to limit ecological processes (e.g., Cushman et al. 2013a).
Meta-replicated landscape genetic studies (e.g., Johnson 2006, independent analysis conducted following the same methodology in more than one area) are necessary to derive reliable inferences about which variables driving gene flow in the study species, and when these features become limiting to gene flow. Despite the fact that pattern–process relationships may differ in different parts of a species’ geographic range, there have been very few spatially replicated landscape genetics studies (Segelbacher et al. 2010; Short Bull et al. 2011; Balkenhol et al. 2015). Thus, it has recently been argued that further advance and generalization of landscape genetics will require a focus on meta-replicated studies to identify limiting factors and thresholds of landscape pattern where they become limiting to gene flow (e.g., Short Bull et al. 2011; Cushman et al. 2013a; Balkenhol et al. 2015).
The stone marten (Martes foina; Erxleben, 1777) is a medium-sized mustelid which is widely distributed throughout much of Europe, central Asia and the Middle East (Tikhonov et al. 2008). In the Iberian Peninsula, the western limit of its distribution, the species is widespread and locally abundant (Reig 2007). The stone marten is considered a food and habitat generalist, but its habitat preferences vary in different parts of its range (Santos and Santos-Reis 2010; Virgós and García 2002). Vergara et al. (2015b) found a tendency of the species to select human associated environments, mostly extensive agricultural areas with a high density of small villages, although in Iberia it is rarely found in or near large urban areas (Basto 2014). The Iberian stone marten prefers forested habitats when the pine marten is absent (Virgós and García 2002), but expresses niche displacement away from preferred pine marten habitats when co-occurring (Vergara et al. 2015b).
Despite the extensive distribution of the stone marten, there have been few population and landscape genetic studies published on the species, in contrast to other species of the genus Martes which have been more thoroughly studied (e.g., pine marten, Ruiz-González et al. 2014, Larroque et al. 2015; American marten, Wasserman et al. 2010, and Fisher, Hapeman et al. 2011). To our knowledge, there have been two landscape genetic studies on the species, which were conducted in Portugal (Basto 2014) and France (Larroque et al. 2016), and two population genetic country level studies, in Bulgaria (Nagai et al. 2012) and the Iberian Peninsula (Vergara et al. 2015a). In Vergara et al. (2015a), the authors combined mitochondrial DNA sequencing and microsatellite genotyping to infer the population genetic structure of the species across Iberia using several Bayesian individual-based clustering approaches and concluded that the main watercourses may act as semi-permeable barriers to gene flow in stone martens. In this paper we used a subset of the genetic data (microsatellites) produced in Vergara et al. (2015a) to investigate the extent to which several landscape variables facilitate or impede stone marten gene flow in each of three study areas which are widely distributed across the Iberian Peninsula and differ substantially in topography, landcover and road density. A main focus of this paper is to evaluate the generality of which factors affect gene flow and to evaluate if factors observed to limit gene flow follow the predictions of Short Bull et al. (2011) and Cushman et al. (2013a, b) who argued that landscape features will only be detected as limiting gene flow when they are highly variable across the landscape such that their pattern directly affects dispersal across a large portion of the population.
Methods
Study areas
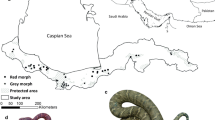
We selected three study areas that were widely distributed across the Iberian Peninsula and exhibited strongly divergent topography, land-cover, and road density (Fig. 1; Table S1). The first study area (BC) covered 15,600 km2 and included most of the Basque Country and part of the adjacent territories of Cantabria, La Rioja and Navarre, ranging from the sea level to 1748 m of the Cantabrian Mountains. The second study area (CAT), of 55,390 km2 in extent, was located in Catalonia and Aragón, with the highest elevation found in the Pyrenees (3400 m). Lastly, we included a region of southern Portugal (SP), of 17,460 km2 in extent, centered on the watersheds of the Tagus and Guadiana Rivers (Fig. 1).
Location of the three study areas (BC, CAT and SP) and the 179 genotyped stone martens (blue, green and purple dots, respectively) in Iberia. Each landcover type is shaded in a different color while the updated distribution of the species in Spain is reported in 10 × 10 km grids (Reig 2007). (Color figure online)
Stone marten genetic diversity and pairwise genetic distances
In this study we used genetic information from 23 microsatellites across 179 individual stone martens across the 3 study areas (BC = 65 individuals, CAT = 60 individuals, SP = 54 individuals), which is a subset of the 333 individuals genotypes for an Iberia-wide population genetic study (Fig. 1; Table S2; Vergara et al. 2015a). Samples were obtained from museum collections, wildlife rescue centers, road-killed individuals and hair samples from live-trapped individuals. Samples were individually genotyped with a novel multiplex panel of 23 autosomal microsatellite markers: 13 species-specific microsatellites and 10 additional markers described in closely related mustelids (Vergara et al. 2015a).
We used FSTAT v.2.9.3 (Goudet 1995) to calculate the Fst values (Weir and Cockerham 1984) of the 179 individuals. The observed and expected heterozygosities (Ho and He) were calculated in GenAlEx 6.5 (Peakall and Smouse 2012). Mean allelic richness across loci (AR) was calculated with the PopGenReport package in R (Adamack and Gruber 2014). Inter-individual GDs were estimated using Rousset’s ar distance (2000) in SPAGEDI v.1.4 (Hardy and Vekemans 2002), which has previously been successfully applied to infer the effect of landscape on genetic structure of continuously distributed carnivores (Ruiz-González et al. 2014; Larroque et al. 2015). We performed a spatial autocorrelation analysis in GenAlEx 6.5 (Peakall and Smouse 2012) to evaluate the null hypothesis of random genetic structure within each area. The autocorrelation coefficient (r) was calculated for each pair of individuals and divided into even distance classes for each of the three areas while the overall significance was evaluated with the heterogeneity test (Peakall and Smouse 2012). Additionally, we tested for the presence of IBD in each of the study areas using a simple Mantel test between the matrix of GDs and the matrix of geographic distances with the adegenet package in R (Jombart 2008).
Predictor variables and parameter space for optimization
We hypothesized a priori that the genetic differentiation of the stone marten could be driven by landscape resistance (IBR) due to four landscape variables: landcover type, elevation, topographical roughness (ruggedness) and/or roads. For each variable we specified a range of maximum resistance value (RMax) and functional shape (power exponent) to define a parameter space for optimizing landscape resistance (e.g., Shirk et al. 2010).
We obtained landcover data from The European landscape database (EEA, CORINE) and we reclassified into nine broad landcover types; natural forests, forestry plantations, agroforestry mosaics, scrublands, pastures, crops, rocks, urban areas and wetlands (Fig. S1). We specified a range of power functions that affect the functional response shape of landscape resistance to landcover type. Following Shirk et al. (2010) we ordered landcover classes from least resistant to most resistant, based on expert knowledge of the species’ ecology, setting resistance of natural forest to 1 and scaling resistance of the other landcover types to increase as power functions with exponents of 0.001, 0.2, 0.4, 0.6, 1, 1.5, 2, 3, and 5 to produce a range of functional shapes ranging from highly concave to highly convex. We also specified five maximum resistance values (10, 20, 30, 40, 50) which were applied to each variable. In combination, this produced 45 (9 power functions * 5 RMax values) resistance parameter combinations for landcover (Fig. S1).
Elevation data was obtained from a 25 m resolution Spanish digital elevation model (Spanish Geographical National Institute; CNIG 2008) and 50 m resolution Portuguese digital elevation model (Portuguese Geographical National Institute). We evaluated a combination of 9 functional shapes of how elevation could affect landscape resistance across 5 levels of RMax, producing 45 combinations of elevation landscape resistance parameterizations (Fig. S1). Specifically, the nine functional shapes of landscape resistance due to elevation was then modeled by applying inverted Gaussian functions, as in Cushman et al. (2006), to produce unimodal functions specifying optimal elevation and rate of increase in resistance as one moves away from that optimal elevation. Specifically, we evaluated three optimal elevations where resistance was set to 1 (100, 500, 900 m) and three standard deviations of the Gaussian function (100, 200, 300 m). The five levels of RMax were 10, 20, 30, 40, 50, as in the landcover parameterization.
Topographic roughness or ruggedness expresses the amount of elevation difference between adjacent cells of a digital elevation grid, providing an objective quantitative measure of topographic heterogeneity (Riley et al. 1999). Roughness was calculated from the elevation layers using the surface gradient and geomorphometric modeling tool (Evans et al. 2014) in ARCGIS. Similarly to landcover, we produced 45 resistance layers for roughness, across the same 9 power function exponents and same 5 RMax values, for each area.
Finally, we modeled resistance arising from roads by building 36 resistance layers (Fig. S1) through the combination of 6 values for each road type (primary roads = 10, 20, 40, 80, 160, 320, secondary roads = 5, 10, 20, 40, 80, 160) based on traffic volume (global roads open access data set).
Multivariate restricted optimization and reciprocal causal modeling
We evaluated the large pool of alternative hypotheses described by the full combination of functional shape and maximum resistance for each of the four predictor variables across each of the three study areas, and optimized the functional form and relative resistance level for each factor. In each study area, we used the multivariate restricted optimization approach developed Shirk et al. (2010) within the reciprocal causal modeling framework (Cushman et al. 2013b) to optimize the relative influence and functional form of the relationship between gene flow and landcover, roads, elevation and topographical roughness. This approach has been shown to reduce Type I error rates in landscape genetic analyses when compared to the original causal modeling approach (Cushman et al. 2014), and has been shown through simulation modeling to correctly identify the landscape factors driving gene flow and reject highly correlated alternative models (e.g., Castillo et al. 2014).
Measures of resistance distance (RD) between sampling points were calculated with the least-cost path approach using the UNICOR software (Landguth et al. 2012). To assess the relationship between GD and RD matrices, we used the partial Mantel test (1967) implemented in the ECODIST package (Goslee and Urban 2007) in R.
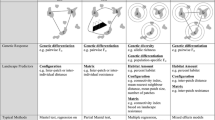
The multivariate restricted optimization approach is conducted in three steps. First, each candidate variable is optimized univariately across a range of functional shapes and maximum resistance values. Second, we conducted a multivariate restricted optimization in which each variable was evaluated across all combinations of its potential parameter values, holding the others at their previously optimal states, and cycling through until the predicted parameter values stabilized for each variable in the context of the optimized values of the other variables. Third, we conducted an analysis of all combinations of variables at their optimal parameter values to determine which variables to include in the final optimized model.
In each of these three optimization steps we used reciprocal causal modeling to simultaneously compete all alternative hypotheses against each other with partial Mantel tests, with the goal of identifying, in each step, the model that was uniquely supported relative to the others. To do this, we calculated a matrix of RS by taking the difference between (a) the partial Mantel r of each candidate model partialling out each alternative model, and (b) the partial Mantel test of the alternative model partialling out the candidate model. A fully supported model would have positive values of (a)–(b) for all alternative models, while no alternative models would have positive values of (a)–(b) when partialling out the fully supported model (Cushman et al. 2013b).
Univariate optimization
For each area, we conducted a univariate optimization (e.g., Shirk et al. 2010) in which we directly competed all functional forms for each individual variable against each other in a reciprocal causal modeling framework (see Fig. S2 for the graphic representation of the univariate optimization step). For each landscape variable in each study area, the best candidate univariate model was the one with positive RS in every comparison (Cushman et al. 2013b) and for which no other model had positive RS when partialling it out. When there was not clear unique support for a single model, we evaluated as potential explanation only those hypotheses that had all positive values in their column. When there were multiple hypotheses with all positive values in their column of the RS matrix, we chose the hypotheses with the lowest number of positive values in its row, to choose the model that was fully supported on test I (focal model partialling out alternative models; supported independently of all other models) and which had the fewest spurious correlations on test II (alternative model partialling out focal model; fewest models supported independently of it; Cushman et al. 2013b).
Multivariate optimization
We used the multivariate optimization approach developed by Shirk et al. (2010) as modified by Castillo et al. (2014). Specifically, we repeated the reciprocal causal modeling for the first input variable, holding all other variables constant at their univariate optima, and combining them through addition. We then changed the model for that first input variable to the one identified in this reciprocal causal modeling step, and repeated for the next predictor variable. This process was repeated until the supported model did not change (Shirk et al. 2010; Cushman et al. 2013b) (see Fig. S3 for the graphic representation of the multivariate optimization step).
Final multivariate combination analysis
In its original form, the Shirk et al. (2010) multivariate optimization assumes that all variables combine at their optimized values to produce the best model of landscape resistance. However, the best model predicting gene flow may not include all predictor variables, even at their optimal form (i.e., the best relationship between landscape structure and gene flow may only be a function of a subset of the predictors; Cushman et al. 2014). We evaluated this by conducting a final reciprocal causal modeling approach between all 15 combinations of the 4 input variables landcover, elevation, roughness and roads (at their multivariate optimization parameters of functional shape and RMax), and identifying, for each study area, which combined model was most supported relative to the others. See Fig. S4 for the graphic representation of the final step of the multivariate optimization.
Finally, we tested whether this final combined model was supported independently of IBD using a final set of partial Mantel tests. For each area, the best multivariate model needed to pass two causal modeling criteria to be significantly better than IBD. First, the partial Mantel correlation between GD and RD of the best multivariate model after partialling out IBD must be significant. Second, the partial Mantel correlation between GD and IBD after partialling out the RD of the best model must be non-significant (Wasserman et al. 2010; Castillo et al. 2014).
Limiting factors
We hypothesized that the effect of a given landscape variable (i.e., landcover, elevation, roughness and roads) on stone marten gene flow would only be detected when it is limiting in the study area (Short Bull et al. 2011; Cushman et al. 2013a). Following Cushman et al. (2013a) we hypothesized that a variable would only be limiting when it was highly variable across the study area such that its heterogeneity affected the gene flow of a large portion of the marten population. Thus, the differences in the supported combined resistance model among study areas may be predictable based on the heterogeneity of the pattern of each predictor variable among study areas. Thus, if one variable is not included in the best supported model, it does not necessarily mean that the variable is not important to stone marten ecology; it may imply that it is not limiting in that particular landscape or it may reflect collinearity with other predictors, overfitting or insufficient statistical power to detect its influence.
We expected landcover to be identified as a landscape factor influencing gene flow in heterogeneous and highly patchy landscapes but not in highly homogenous ones (e.g., Short Bull et al. 2011, Cushman et al. 2013b). Thus, we used FRAGSTATS software v 4.2 (McGarigal et al. 2012) to calculate five landscape level metrics [patch density (PD), edge density (ED), correlation length (GYRATE_AM), Shannon’s diversity (SHDI) and aggregation index (AI)] that have previously been shown to measure attributes of landscape heterogeneity that are highly sensitive measures of when landscape patterns limit gene flow processes (e.g., Cushman et al. 2013b). PD is the density of patches of all cover types in the landscape mosaic, ED is the length of edges between dissimilar patch types per unit area, correlation length is the area-weighted mean radius of patch gyration, and is the expected value of the distance one can traverse the landscape when starting in a random location and moving in a random direction before leaving the patch of origin. Shannon’s diversity is the diversity of patch types, combining both richness and evenness as formulated in the Shannon diversity index. AI is a measure of landscape heterogeneity, where high levels of aggregation occur when a large proportion of cells in the landscape are surrounded by cells of the same cover class.
Similarly, we expected elevation to be identified as influencing gene flow in study areas with a relatively high variance of elevation, but not in those where the topography is relatively flat (e.g., Short Bull et al. 2011). Thus, to quantify the heterogeneity of the landscape in terms of its elevation we calculated the mean and the standard deviation of elevation in ARCGIS.
Likewise, topographical roughness is likely to influence gene flow in study areas with a high degree of ruggedness, while in flatter study areas topography would not limit gene flow and thus would not influence population structure. We calculated the mean of the topographical roughness surface (Evans et al. 2014) within a 1000 m focal radius in ARCGIS. Finally, the road network is expected to limit gene flow only in areas where roads are extensive and dissect the landscape, but not where roads are few and do not fragment the area (e.g., Short Bull et al. 2011). To measure the extensiveness of roads across the landscape we calculated the correlation length of roads in FRAGSTATS (class-level metric correlation length; GYRATE_AM), treating roads as a categorical cover type. Correlation length provides a measure of landscape connectivity and represents the average traversability of the landscape for an organism that is confined to remain within a single patch.
Results
Genetic diversity
The CAT area had the highest genetic diversity (AR = 4.971 ± 1.733, Ho = 0.530 ± 0.035, He = 0.611 ± 0.031) followed by BC (AR = 4.414 ± 1.967, Ho = 0.523 ± 0.027, He = 0.584 ± 0.031), with SP lowest (AR = 3.733 ± 1.517, Ho = 0.474 ± 0.035, He = 0.521 ± 0.035). Fst values were highest between SP and CAT (Fst = 0.1148) and SP and BC (Fst = 0.1082), and lowest between BC and CAT (Fst = 0.0541), consistent with the geographic distance among them. All three correlograms (e.g., one for each study area) were significant (heterogeneity test p < 0.01) and showed a pattern of decreasing relatedness with increasing distance (Fig. 2). The x-intercept provides an estimate of the extent of non-random genetic structure, and varied from 50.4 km in BC and 59.1 km in SP to 93.8 km in CAT. The genetic versus geographic distance scatterplot provides a means to compare the relationship between geographic and GD in all three study areas at the same scale (Fig. 2). The CAT study area showed low genetic structure and the highest intercept, when comparing among areas (Fig. 2d).
a–c Spatial autocorrelation analysis. Correlogram plots of the autocorrelation coefficient (r) as a function of 10 distance classes for BC and SP areas and 30 distance classes for CAT area calculated in GenAlEx. d Scatterplot of the geographic distance (Log km) versus genetic distance (Rousset’s ar distance, dots) and linear fit (lines) for BC, SP and CAT in blue, purple and green, respectively (Fig. 1). (Color figure online)
Optimized resistance models
The best supported univariate parameterization differed among study areas for each of the four predictor variables (Table 1). Optimized univariate models for landcover had the lowest maximum resistance values in BC and SP (e.g., 10) but the highest in CAT (e.g., 50), and presented high variation in power function exponents (0.001–5; Fig. S1). In CAT and SP the landcover model identified in univariate and multivariate optimization steps was the same, while in BC the power function exponent decreased slightly in the multivariate optimization (Fig. S5).
When modeling IBR arising from elevation, the best supported univariate and multivariate models all had an optimal elevation of 500 m (Fig. S6). However, across the three study areas, all the three standard deviations and the maximum and minimum resistance values were selected. The elevation model identified by univariate and multivariate optimization was the same in CAT but varied in BC and SP.
Univariate and multivariate models for roughness did not change between the univariate and multivariate optimizations in CAT, but the power exponent varied slightly in BC and also in SP, both at the maximum RMax level of 50 (Fig. S7). Lastly, among the road resistance models, BC showed a low resistance to primary and secondary roads in univariate and multivariate optimization and so did CAT in the univariate model selection step. On the other hand, in the multivariate models for the SP and CAT study areas the influence of primary roads was relatively weak, as measured by RMax, while the influence of the secondary roads was high (Fig. S8). The movement of the supported model across the univariate and multivariate optimizations in the hypothesis space is reported in Fig. S9.
Final model selection
The final multivariate combination analysis identified landcover (L), elevation (E) and topographical roughness (R) (LER) as the best IBR model predicting gene flow in BC (Fig. 3). The best supported IBR model for CAT was LERRd, which included all the four variables [e.g., LER and roads (Rd)], while the best IBR model for SP included only topographical variables (e.g., E and R; Fig. 3). Thus, the three study areas each had a different most-supported IBR model, but elevation and topographic roughness were landscape features identified as influencing gene flow in all of them.
Final reciprocal causal modeling matrix for the 15 multivariate combination analyses performed (including all combinations of the 4 input variables; landcover, elevation, roughness and roads, at their optimal form). Columns indicate focal models, and rows indicate alternative models. The color gradient from blue to yellow indicates relative support for the focal model independent of the alternative model (e.g., focal model | alternative model − alternative model | focal model is positive). The best IBR model predicting gene flow in each study area; a BC, b CAT and c SP are indicated in black. (Color figure online)
IBD was statistically significant in all areas (Mantel r values of BC = 0.233, CAT = 0.161, SP = 0.237; p < 0.001) based on a simple Mantel test (Fig. S10). The Mantel regression plot displayed a single high-density nucleus (Fig. S10). The final reciprocal causal modeling showed IBR independently of IBD in the BC and SP study areas (e.g., the Mantel r value for IBR partialling out IBD is positive and the Mantel r value for IBD partialling out IBR is negative) showing that the genetic differentiation among stone martens in these areas is driven by IBR independently of IBD (Table 2). However, the IBR model for CAT (LERRd) was not supported independently of IBD (e.g., Mantel values for IBR partialling out IBD and IBD partialling out IBR are both positive; Table 2).
Limiting factors
Landcover was included in the multivariate optimized models in the BC and CAT study areas, and both study areas presented high landscape heterogeneity as measured by five FRAGSTATS landscape-level metrics (Table S1). However, landcover was not among the variables included in the best supported IBR model for SP, which also had substantially lower heterogeneity of the landcover mosaic (Table S1). The BC and CAT study areas had high variation in elevation and roughness, while SP had relatively lower topographical variation. Roads were included only in the optimized CAT model, and the correlation length of roads in CAT area was 176% greater than in BC and 134% greater than in SP area (Table S1).
Discussion
Topography as a common driver of genetic differentiation
To be detected as a factor influencing gene flow a landscape feature must significantly vary across the study area in a way that limits gene flow (Short Bull et al. 2011; Cushman et al. 2013a). In this study, all three replicate landscapes multivariate optimization identified elevation and topographical roughness as drivers of genetic differentiation. This is interesting given that SP has a substantially lower degree of topographical complexity that do CAT or BC. This suggests that elevation and topographical roughness affect gene flow of stone martens even at the relatively lower heterogeneity seen in SP (Table S1). Stone marten gene flow presented a consistent response to topography across the three study areas, with optimal elevation of 500 m and nonlinearly increasing resistance with increasing topographical roughness. This suggests that stone marten dispersal movement may be highly related to extent of suitable climatic zones (with elevation as a proxy variable), as was also found for American marten by Wasserman et al. (2010, 2012), or to changes in vegetation types with elevation. In addition, the observation of a consistent response to topographical roughness suggests that cliffs, ridges and steep mountains may act as filters or partial barriers to gene flow of this species.
Variation in landscape resistance hypotheses among areas
In contrast to their common response to elevation and topographical roughness, the three study areas differed with regard to the other variables (roads and landuse). In SP the genetic differentiation of the stone marten was a function of elevation and roughness alone, while landcover and roads were not included in the best supported resistance models. This is consistent with our a priori expectations given that SP had the lowest heterogeneity of landcover and lowest correlation length of roads of the three study areas, suggesting these variables are not variable enough in SP to limit gene flow. Consistent with our results, Basto (2014) found that landcover did not have a clear influence on the genetic structure of the stone marten in Portugal, but suggested a restricted gene flow across rivers (e.g., the most resistant landcover class). However, we did not find a clear response to rivers is SP, where landcover was not included in the best supported resistance model.
In addition to topographic variables, the BC study area also included landcover in its final optimized model of landscape resistance to stone marten gene flow. We expected land cover to be limiting in BC a priori given its very high landscape heterogeneity. Cushman et al. (2012) found that the strength of spatial genetic structure is related to the heterogeneity of the landscape mosaic and the degree of contrast in resistance to gene flow among the different landcover types. Correlation length (GYRATE_AM) and PD are among the most effective landscape metrics for predicting the effects of landscape heterogeneity on genetic differentiation (Cushman et al. 2012). Therefore, based on the comparison of the values obtained in each of the study areas, we expected the BC study area to show the strongest relationship between gene flow and landcover because it had lowest GYRATE_AM and highest PD. Consistent with this a priori expectation, the best supported model in BC area (LER), which was supported independently of IBD, suggested that landcover influences the genetic differentiation among individuals in this area. However, the final multivariate parameterization of resistance due to landcover in the BC study area indicated that all nine landcover types had very similar resistance coefficients, suggesting little difference among landcover types in their resistance to gene flow. Thus, while our results identify a significant effect of landcover, as expected due to the heterogeneity of the BC area, they also suggest that gene flow in BC is primarily limited by topographical variables, with relatively weaker effects of landcover.
It should be noted that the ranking of land-cover classes in order of increasing landscape resistance to the stone marten gene flow was based on expert knowledge of the species, and it would have been computationally infeasible to run the optimization across all orders of possible resistance ranking as well as functional shape and maximum resistance. However, a misspecification of the order of landcover classes in terms of their relative resistance could mask the influence of landcover in the genetic structuring of the species. Future research should empirically evaluate a full range of landscape resistance parameterization for landcover, including alternative relative resistance ranks of landcover classes.
The best supported IBR hypothesis for the CAT study area included all four of the predictor variables, as we would expect based on limiting factors. The CAT landscape is highly heterogeneous, with high landscape fragmentation and pronounced topographic variation. The forth variable, roads, was only identified as influencing gene flow in CAT, which is consistent with the high correlation length of roads in this area. This suggests that the road network may only impede stone marten gene flow when it exceeds a threshold of extensiveness where it begins to limit dispersal, below which it is not limiting to gene flow (Short Bull et al. 2011; Cushman et al. 2013a).
Several factors may explain the fact that the best supported IBR hypothesis (e.g., LERRd) was not supported independently of IBD in CAT, the largest study area. First, CAT had the highest genetic diversity of all areas (e.g., number of alleles and mean AR; Table S1), suggesting a greater population size, which could result in high gene flow and low genetic drift and thus, could limit the emergence of genetic structure due to either IBR or IBD. Second, to obtain a higher number of individuals and coverage, museum samples were included in CAT area (a quarter of all samples were collected before 2006). Thus, the large temporal variation in sampling periods among recent (2006–2012) and older samples could hinder the identification of the landscape variables limiting gene flow in CAT area. Third, the independent IBD signal could be an indicator of additional spatial variance in genetic differentiation, not explained by the IBR model because it does not perfectly reflect the true process driving gene flow. Nevertheless all four factors (LERRd) are supported and appear limiting as they are all included in the top IBR model, and even if not independent of IBD, the IBR model was more strongly supported than IBD (e.g., larger partial Mantel r in reciprocal causal modeling matrix).
Differences in spatial and temporal scales among areas
Several studies have underlined the importance of the spatial and temporal scales of the sampling design when conducting a landscape genetic study (Cushman and Landguth 2010; Segelbacher et al. 2010; Oyler-McCance et al. 2013). The spatial scale (i.e., sampling design and extent of the study area) is known to influence the ability to correctly infer the impacts of landscape patterns on gene flow (Anderson et al. 2010; Oyler-McCance et al. 2013). In this study, however, samples were opportunistically collected and therefore, each of the three study areas presents a particular sampling scheme; while in CAT area samples are quite homogeneously distributed, in SP area most of the samples are concentrated in the southwest. In addition, the CAT area is three times larger than BC or SP areas, which can influence the power of the model to detect the underlying spatial genetic structure (Cushman and Landguth 2010; Oyler-McCance et al. 2013).
The temporal scale can also affect the power of landscape genetics methods to detect spatial genetic structure and landscape correlates of genetic differentiation (Anderson et al. 2010; Cushman and Landguth 2010). In BC and SP areas >85% of the samples were collected between 2006 and 2012 but in CAT area, in order to obtain a higher spatial coverage, approximately a 25% of the samples were museum specimens (i.e., collected before 2006). Thus the temporal variation in sampling periods, which differed among areas, could have influenced the ability to make inferences on stone marten gene flow based on the observed current spatial genetic structure.
Importance of meta-replication studies
McGarigal and Cushman (2002) argued that landscape-level replication was the most important characteristic for studies to produce reliable and generalizable inferences about the effects of habitat heterogeneity on populations and species distributions. For the same reason, meta-replication of landscape studies is critical to understand the factors that control gene flow. Balkenhol et al. (2015) identified meta-replicated landscape genetic studies as one of the areas of highest priority for future research. Short Bull et al. (2011) conducted one of the first meta-replicated landscape genetic studies, and their results motivated Cushman et al. (2013a) to use simulation modeling to understand limiting factors and how they act in different study areas, and the thresholds where they emerge. These studies demonstrated that differences in supported models may arise when landscape features do not limit gene flow in a given landscape, because of their extent or pattern, but do limit gene flow in other landscapes (Short Bull et al. 2011; Cushman et al. 2013a).
Our results demonstrate that key variables explaining the stone marten gene flow differed among areas in ways consistent with our a priori expectations based on limiting factors. These results confirmed the importance of meta-replicated studies to draw robust and general conclusions regarding a species-specific response to landscape characteristics. However, to date, few landscape genetic studies include spatial replicates (e.g., Short Bull et al. 2011; Larroque et al. 2015), highlighting the urgent need for more studies of this kind to understand limiting factors across multiple landscapes (Balkenhol et al. 2015).
In addition to the importance of meta-replication in general, it is critical for future studies to undertake meta-replicated analyses with high levels of spatial replication across controlled gradients of landscape structure in a comparative mensurative replicated design (e.g., McGarigal and Cushman 2002). Only then will there be sufficient power to generalize results and rigorously identify thresholds where heterogeneity and resistance contrast are sufficient to limit gene flow. Based on past empirical (Short Bull et al. 2011) and simulation (Cushman et al. 2013b) studies, we were able to propose a priori hypotheses about which landscapes we would expect each predictor variable to be limiting to gene flow and our results were consistent with those a priori expectations. We hope future studies will continue to explore limiting factors in well replicated comparative mensurative studies that a priori select replicate landscapes across a range of landscape heterogeneities for multiple predictor variables.
Conclusions
The main conclusions of this study are that roads, landcover and topography are factors that can limit stone marten gene flow. Our results suggest that roads will limit gene flow when roads are extensive and fragment the study area, as was also found by Short Bull et al. (2011) for American black bear. We found that topography can limit gene flow when the landscape has high roughness and high variation in elevation, as was seen in Short Bull et al. (2011), and Wasserman et al. (2010) for American marten. Landcover type can be limiting to stone marten gene flow when the landscape is highly heterogeneous, with low resistance cover types distributed in a patchy and irregular pattern interspersed with high contrast cover types, as was also seen by Short Bull et al. (2011). Our results support the idea that topography, landcover and roads can limit stone marten gene flow and identify some circumstances where they do or do not limit gene flow in several case-study landscapes.
References
Adamack AT, Gruber B (2014) PopGenReport: simplifying basic population genetic analyses in R. Methods Ecol Evol 5:384–387
Anderson CD, Epperson BK, Fortin MJ, Holderegger R, James PMA, Rosenberg MS, Scribner KT, Spear S (2010) Considering spatial and temporal scale in landscape-genetic studies of gene flow. Mol Ecol 19:3565–3575
Balkenhol N, Cushman SA, Waits LP, Storfer A (2015) Current status, future opportunities, and remaining challenges in landscape genetics. In: Balkenhol N, Cushman SA, Waits LP, Storfer A (eds) Landscape genetics: concepts, methods, applications. Wiley, Chichester, pp 247–256
Balkenhol N, Gugerli F, Cushman SA, Waits LP, Coulon A, Arntzen JW, Holderegger R, Wagner HH (2009) Identifying future research needs in landscape genetics: where to from here? Landscape Ecol 24:455–463
Basto MP (2014) Population and landscape genetics of the stone marten and red fox in Portugal: implications for conservation management of common carnivores. MSc Thesis. University of Lisbon
Bradburd GS, Ralph PL, Coop GM (2013) Disentangling the effects of geographic and ecological isolation on genetic differentiation. Evolution 67:3258–3273
Castillo J, Epps CW, Davis AR, Cushman SA (2014) Landscape effects on gene flow for a climate-sensitive montane species, the American pika. Mol Ecol 23:843–856
Cushman SA, Landguth EL (2010) Spurious correlations and inference in landscape genetics. Mol Ecol 19:3592–3602
Cushman SA, Max T, Meneses N, Evans LM, Ferrier S, Honchak B, Whitham TG, Allan GJ (2014) Landscape genetic connectivity in a riparian foundation tree is jointly driven by climatic gradients and river networks. Ecol Appl 24:1000–1014
Cushman SA, McKelvey KS, Hayden J, Schwartz MK (2006) Gene flow in complex landscapes: testing multiple hypotheses with causal modeling. Am Nat 168:486–499
Cushman SA, McRae BR, McGarigal K (2015) Basics of landscape ecology: an introduction to landscapes and population processes for landscape geneticists. In: Balkenhol N, Cushman SA, Waits LP, Storfer A (eds) Landscape genetics: concepts, methods, applications. Wiley, Chichester, pp 11–34
Cushman SA, Shirk A, Landguth EL (2012) Separating the effects of habitat area, fragmentation and matrix resistance on genetic differentiation in complex landscapes. Landscape Ecol 27:369–380
Cushman SA, Shirk A, Landguth E (2013a) Landscape genetics and limiting factors. Conserv Genet 14:263–274
Cushman SA, Wasserman T, Landguth E, Shirk A (2013b) Re-evaluating causal modeling with Mantel tests in landscape genetics. Diversity 5:51–72
European Environmental Agency. CORINE land cover. http://www.eea.europa.eu/data-and-maps/data/corine-land-cover-2006-raster-2
Evans J, Oakleaf J, Cushman SA, Theobald D (2014) An ArcGIS toolbox for surface gradient and geomorphometric modeling, version 2.0-0
Global roads open access data set, version 1 (gROADSv1). NASA Socioeconomic Data and Applications Center (SEDAC), Palisades. http://sedac.ciesin.columbia.edu/data/set/groads-global-roads-open-access-v1/data-download
Goslee SC, Urban DL (2007) The ecodist package for dissimilarity-based analysis of ecological data. J Stat Softw 22:1–19
Goudet J (1995) FSTAT (version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Graves T, Beier P, Royle JA (2013) Current approaches using genetic distances produce poor estimates of landscape resistance to interindividual dispersal. Mol Ecol 22:3888–3903
Graves T, Wasserman TN, Ribeiro MC, Landguth EL, Spear SF, Balkenhol N, Higgins CB, Fortin MJ, Cushman SA, Waits LP (2012) The influence of landscape characteristics and home-range size on the quantification of landscape-genetics relationships. Landsc Ecol 27:253–266
Guillot G, Rousset F (2011) On the use of the simple and partial Mantel tests in presence of spatial auto-correlation. Syst Biol 2011:1–13
Guillot G, Rousset F (2013) Dismantling the Mantel tests. Methods Ecol Evol 4:336–344
Hapeman P, Latch EK, Fike J, Rhodes OE, Kilpatrick CW (2011) Landscape genetics of fishers (Martes pennanti) in the Northeast: dispersal barriers and historical influences. J Hered 102:251–259
Hardy O, Vekemans X (2002) SPAGEDI: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hargrove WW, Pickering J (1992) Pseudoreplication: a sine qua non for regional ecology. Landscape Ecol 6:251–258
Johnson DH (2006) The many faces of replication. Crop Sci 46:2486–2491
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Landguth EL, Cushman SA, Schwartz M, McKelvey K, Murphy M, Luikart G (2010) Quantifying the lag time to detect barriers in landscape genetics. Mol Ecol 19:4179–4191
Landguth EL, Hand BK, Glassy J, Cushman SA, Sawaya MA (2012) UNICOR: a species connectivity and corridor network simulator. Ecography 35:9–14
Larroque J, Ruette S, Vandel JM, Devillard S (2015) Divergent landscape effects on genetic differentiation in two populations of the European pine marten (Martes martes). Landscape Ecol 31:517
Larroque J, Ruette S, Vandel J-M, Queney G, Devillard S (2016) Age and sex-dependent effects of landscape cover and trapping on the spatial genetic structure of the stone marten (Martes foina). Conserv Genet 17:1293–1306
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Manel S, Schwartz MK, Luikart G, Taberlet P (2003) Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol 18:189–197
McGarigal K, Cushman SA (2002) Comparative evaluation of experimental approaches to the study of habitat fragmentation effects. Ecol Appl 12:335–345
McGarigal K, Cushman SA, Ene E (2012) FRAGSTATS v4: spatial pattern analysis program for categorical and continuous maps. Computer software program produced by the authors at the University of Massachusetts, Amherst. http://www.umass.edu/landeco/research/fragstats/fragstats.html
Nagai T, Raichev E, Tsunoda H (2012) Preliminary study on microsatellite and mitochondrial DNA variation of the stone marten Martes foina in Bulgaria. Mamm Study 37:353–358
Oyler-McCance SJ, Fedy BC, Landguth EL (2013) Sample design effects in landscape genetics. Conserv Genet 14:275–285
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Portuguese Geographical National Institute. Portuguese digital elevation model. 50 m resolution
Radford JQ, Bennett AF, Cheers GJ (2005) Landscape-level thresholds of habitat cover for woodland-dependent birds. Biol Conserv 124:317–337
Reig S (2007) Martes foina Erxleben, 1777. In: Palomo L, Gisbert J, Blanco J (eds) Atlas y Libro Rojo de los Mamíferos Terrestres de España. Dirección General de Biodiversidad-SECEM-SECEMU, Madrid, pp 305–307
Riley SJ, DeGloria SD, Elliot R (1999) A terrain ruggedness index that quantifies topographic heterogeneity. Intermt J Sci 5:1–4
Rousset F (2000) Genetic differentiation between individuals. J Evol Biol 13:58–62
Ruiz-González A, Gurrutxaga M, Cushman SA, Randi E, Gómez-Moliner B (2014) Landscape genetics for the empirical assessment of resistance surfaces: the European pine marten (Martes martes) as a target-species of a regional ecological network. PLoS ONE 9:19
Santos MJ, Santos-Reis M (2010) Stone marten (Martes foina) habitat in a Mediterranean ecosystem: effects of scale, sex, and interspecific interactions. Eur J Wildl Res 56:275–286
Schwartz MK, McKelvey KS (2009) Why sampling scheme matters: the effect of sampling scheme on landscape genetic results. Conserv Genet 10:441–452
Segelbacher G, Cushman SA, Epperson BK, Fortin MJ, Francois O, Hardy OJ, Holderegger R, Taberlet P, Waits LP, Manel S (2010) Applications of landscape genetics in conservation biology: concepts and challenges. Conserv Genet 11:375–385
Shirk AJ, Landguth EL, Cushman SA (2017) A comparison of regression methods for model selection in individual-based landscape genetic analysis. Mol Ecol Res (in review)
Shirk AJ, Raphael MG, Cushman SA (2014) Spatiotemporal variation in resource selection: insights from the American marten (Martes americana). Ecol Appl 24:1434–1444
Shirk AJ, Wallin DO, Cushman SA, Rice CG, Warheit KI (2010) Inferring landscape effects on gene flow: a new model selection framework. Mol Ecol 19:3603–3619
Short Bull RA, Cushman SA, Mace R, Chilton T, Kendall KC, Landguth EL, Schwartz MK, McKelvey K, Allendorf FW, Luikart G (2011) Why replication is important in landscape genetics: American black bear in the Rocky Mountains. Mol Ecol 20:1092–1107
Spanish Geographical National Institute, CNIG (2008) Spanish digital elevation model. 25 m resolution
Tikhonov A, Cavallini P, Maran T, Krantz A, Herrero J, Giannatos G, Stubbe M, Libois R, Fernandes M, Yonzon P, Choudhury A, Abramov A, Wozencraft C (2008) Martes foina. IUCN 2013. IUCN Red List of Threatened Species. Version 2013.2. www.iucnredlist.org. Accessed 5 Sep 2015
Turner MG, Gardner RH (1991) Quantitative methods in landscape ecology. The analysis and interpretation of landscape heterogeneity. In: Ecological studies, vol 82. Springer, New York
Vergara M, Basto M, Madeira M, Gómez-Moliner B, Santos-Reis M, Fernandes C, Ruiz-González A (2015a) Inferring population genetic structure in widely and continuously distributed carnivores: the stone marten (Martes foina) as a case study. PLoS ONE 10(7):e0134257
Vergara M, Cushman SA, Urra F, Ruiz-González A (2015b) Shaken but not stirred: multiscale habitat suitability modeling of sympatric marten species (Martes martes and Martes foina) in the northern Iberian Peninsula. Landscape Ecol 31:1241–1260
Virgós E, García FJ (2002) Patch occupancy by stone martens Martes foina in fragmented landscapes of central Spain: the role of fragment size, isolation and habitat structure. Acta Oecol 23:231–237
Wasserman TN, Cushman SA, Schwartz MK, Wallin DO (2010) Spatial scaling and multi-model inference in landscape genetics: Martes americana in northern Idaho. Landscape Ecol 25:1601–1612
Weir B, Cockerham C (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Wiens JA, Chr N, Van Horne B, Ims RA (1993) Ecological mechanisms and landscape ecology. Oikos 66:369–380
Zeller KA, Creech TG, Millette KL, Crowhurst RS, Long RA, Wagner HH, Balkenhol N, Landguth EL (2016) Using simulations to evaluate Mantel-based methods for assessing landscape resistance to gene flow. Ecol Evol 6:4115–4128
Acknowledgments
MV (Ref: RBFI-2012-446) and ARG (Ref: DKR-2012-64) were supported by a PhD and Post-doctoral Fellowships awarded by the Department of Education, Universities and Research of the Basque Government. US Forest Service Rocky Mountain Research Station supported SC’s work on this Project. The authors wish to thank all the people and institutions for supplying some of the tissue samples used in this study and all the people directly involved in the collection of samples, including rangers, vets and field researchers (all listed in Vergara et al. 2015a, b). Special thanks to Mafalda P. Basto, Margarida Santos-Reis and Carlos Fernandes, researchers from CE3C—Centre for Ecology, Evolution and Environmental Changes, Faculdade de Ciências, Universidade de Lisboa, for their previous collaboration in stone marten genetics in the Iberian Peninsula (Vergara et al. 2015a, b).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vergara, M., Cushman, S.A. & Ruiz-González, A. Ecological differences and limiting factors in different regional contexts: landscape genetics of the stone marten in the Iberian Peninsula. Landscape Ecol 32, 1269–1283 (2017). https://doi.org/10.1007/s10980-017-0512-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10980-017-0512-0