Abstract
Context
Habitat fragmentation is likely to have deleterious genetic consequences for plant populations. Although the genetic effects of fragmentation in plants have been investigated in various landscapes, such studies are scarce in urban landscapes where forests tend to be fragmented and have a complex internal structure.
Objectives
This study aimed to determine the factors, including patch and sub-patch level spatial factors, affecting the genetic diversity of a herbaceous species in urban fragmented forests.
Methods
We collected 30–39 leaf samples of Viola grypoceras A. Gray var. grypoceras, a perennial herbaceous species with short-distance seed dispersal, from 12 fragmented and 12 suburban forests each at Kyoto City, Japan, and analyzed the genetic diversity of this species by developing six simple sequence repeat markers. Field survey was conducted to collect demographic and spatial data.
Results
There was no significant difference in allelic richness between the urban fragmented and suburban forests. However, statistical analysis revealed that the area of vegetation, distribution pattern of populations in a forest, and average distance between nearest populations affected the genetic diversity of this species in urban fragmented forests.
Conclusion
Although V. grypoceras has traits that allow it to tolerate fragmentation, such as self-pollination and seed bank-formation ability, pure loss of habitat and reduced fragment size might have deleterious effects on this species, and these effects might become more apparent if fragmentation continues to proceed in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The direct effects of habitat fragmentation include four components: (1) the creation of small patches, (2) alternation of landscape processes, (3) increased spatial isolation of remnant fragments, and (4) the reduction of population sizes (Andren 1994; Hobbs and Yates 2003; Honnay and Jacquemyn 2007). These factors influence the follow-on consequences for ecosystem and species such as altered environmental conditions, edge effects, increased incidence and abundance of invasive species, changed disturbance regimes, altered species interactions, and genetic deterioration (Hobbs and Yates 2003). In particular, increased isolation and reduction of population sizes can result in genetic deterioration on plant populations. Populations in fragmented areas might show genetic erosion, inbreeding depression, and reduced reproductive success because of increased genetic drift and inbreeding, reduced gene flow, and disrupted pollination processes (Young et al. 1996; Kwak et al. 1998; Oostermeijer et al. 2003). Over the long term, fragmentation might also reduce the ability of populations to adapt to changing environments, thereby increasing the extinction events of local populations (Mills and Tallmon 1999). Although the genetic effects of fragmentation in plants have been investigated in various landscapes—grasslands (e.g., Young et al. 1999), forests (e.g., Fore et al. 1992; Young et al. 1993; Cruzan 2001), and agricultural areas (e.g., Berg et al. 1998)—urban landscape where forests tend to be fragmented in small isolated remnants and have a complex internal structure consisting of naturally and artificially covered areas have remained relatively unexplored.
Urban development is occurring on an unprecedented scale (Goddard et al. 2010). Among the many human activities that cause habitat loss, urban development leads to high local extinction rates and frequently eliminates a large majority of native species (McKinney 2002). Generally, forests in urban areas are fragmented into small isolated patches, and are embedded in an inhospital anthropogenic matrix (Fahrig 2003; Alberti 2005). These isolated patches experience strong environmental pressures that are related to the urban context, e.g., recreational activities, ornamental and exotic tree plantations, and increased pollution and eutrophication (Van Rossum 2008). All of these processes can lead to soil degradation, trampling, disturbance, and degradation of herbaceous forest vegetation (Van Rossum 2008). As urbanization extends, the importance of urban forests increases, especially in areas that retain some biodiversity (Honnay et al. 1999; Godefroid and Koedam 2003; Imanishi et al. 2005). To conserve these fragmented habitats, the problem of decreasing genetic diversity in small and isolated populations (Ledig 1992) must also be considered. However, the genetic consequences of habitat fragmentation have been rarely investigated in urban plant populations (Van Rossum 2008).
To our knowledge, only a few studies have investigated the genetic effects of habitat fragmentation on herbaceous species in urban forests. Culley et al. (2007) showed that urban habitat fragmentation did not impede gene flow of Viola pubescens in southwestern Ohio, USA. Horning and Webster (2009) also showed that there was no pronounced effect of habitat fragmentation on the degree of genetic differentiation and subsequent gene flow among remnant Lilium philadelphicum populations in a highly disturbed urban landscape of the Midwest United States. On the other hand, Van Rossum (2008) found that urban population of Primula elatior showed higher genetic erosion in small populations during recruitment. These studies indicate that effects of habitat fragmentation on genetic diversity in urban fragmented forests cannot be generalized, and that the relationship between spatial structure of urban forests and genetic diversity, which is important for conservation planning, needs to be determined. Although previous studies treat each forest as a minimum spatial unit for analysis, a high degree of environmental heterogeneity is usually found in forest habitats (Whigham 2004). In urban forests, artificial alteration such as the creation of pathways tends to increase the heterogeneity. Environmental heterogeneity such as light and moisture conditions influences competition among woodland herbs (Whigham 2004) and determines the spatial distribution of herbaceous plant species within a forest. The resultant distance and distribution pattern of populations in a forest may be an important factor that controls the frequency of gene flow. Thus, we hypothesized that the internal spatial arrangement such as distance and distribution pattern of populations in a forest might also affect the genetic diversity of a herbaceous plant. This study aimed to determine the factors, including spatial factors at patch and sub-patch levels, affecting the genetic diversity of a herbaceous species in urban fragmented forests. The following questions were addressed: (1) Is genetic diversity of herbaceous species in urban fragmented forests lower than that in suburban continuous forests? (2) Does the internal spatial arrangements of populations in a forest significantly affect the genetic diversity of herbaceous species in urban fragmented forests?
Materials and methods
Study species
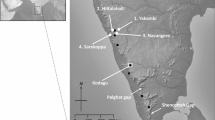
Viola grypoceras A. Gray var. grypoceras is a common violet that is distributed all over Japan (Fig. 1) and grows both in urban fragmented forests and suburban continuous forests. It exhibits dimorphic cleistogamy, producing two types of flowers at different periods of a year: purple chasmogamous flowers are produced first in early spring and inconspicuous cleistogamous flowers appear subsequently in late spring. Individuals continue to produce self-pollinated cleistogamous flowers until plant senescence in late autumn. Chasmogamous flowers are visited by bumblebees, butterflies, and bee flies. Seeds are dispersed ballistically up to 1.5 m away from the maternal plant as well as by ants (Hama 2002). The average distance of seed dispersal is estimated to be less than 3 m a year (Culver and Beattie 1978; Hama 2002).
Study sites
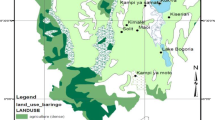
The study sites were selected from a disturbed urban area and non-disturbed suburban mountains in Kyoto City, Japan (Fig. 2). The urban area targeted in this study was developed at least 80 years ago, and many fragmented forests exist in this area. Terra/ASTER satellite image (June 24, 2010; spatial resolution: 15 m) was imported to ArcGIS version 10.1 (Environmental Systems Research Institute, Inc., USA). A normalized differential vegetation index (NDVI) image was derived, and forests were extracted using an NDVI value of greater than or equal to 0.20. Subsequently, 33 fragmented forests larger than 1 ha were selected from the urbanized area as study sites. Additionally, 14 suburban mountainous sites in which V. grypoceras were found in our preliminary survey were also selected as study sites in order to compare genetic diversity between these sites and the urban forests.
Field survey
Field survey was conducted from March to May 2012. Urban fragmented forests were surveyed in all areas except for forbidden grounds. Because the habitats of V. grypoceras in suburban mountainous areas were embedded in large forests, we surveyed habitats of V. grypoceras along the trails. The following data were collected at each study site: number of populations (N P), number of individuals (N I), number of individuals with chasmogamous flowers (N IF), and number of chasmogamous flowers (N F). A population was defined by distance between the individuals: When the average dispersal distance of 3 m was exceeded, the individual was considered to belong to another population. When more than 30 individuals were found at a study site, leaf samples were collected for genetic analysis from 30 to 39 individuals. The number of samples from each study site was selected in view of the number of individuals in that site. Next, the leaf samples were frozen and stored below −30 °C.
Spatial analysis of urban forests
Area of urban forests (A SITE) and distance between urban forests and the nearest suburban mountains (D MT) were derived in ArcGIS in order to analyze their effects on genetic diversity at the patch level or inter-forest level.
At the sub-patch level or intra-forest level, which we defined as a spatial scale that describes factors within a forest, we derived the following variables for each urban forest: area of vegetated area (A VEG) and area of non-vegetated area (A NOVEG) were derived using the Calculate Geometry tool in ArcGIS because urban forests include non-vegetative features such as walking paths and buildings for human use. The observed average distance between the nearest neighbor populations (D O) and the average nearest neighbor z-score (Z ANN) were calculated using the Average Nearest Neighbor tool in ArcGIS. Z ANN is a statistical index that is derived from the observed average distance and expected distance between the nearest neighbor populations under the hypothesis that populations are randomly distributed in a forest (Ebdon 1985). Z ANN indicates a dispersed distribution pattern when it is greater than 1.96, a clustered distribution pattern when it is less than −1.96, and a random distribution pattern when it is greater than −1.96 and less than 1.96, at the 95 % confidence level. As has been described later, we adopted the absolute value of Z ANN (|Z ANN|) for model construction; thus, |Z ANN| of less than 1.96 indicates that the distribution of populations in a forest is closer to a random pattern, that is, between dispersed and clustered patterns. Moreover, the distance between the farthest populations at each forest (D MAX) and standard deviation of distances between observed populations and their geometric mean center (D SD), which indicates the degree of dispersion/compactness around their geometric mean center, were computed using the Proximity Analysis tool and Standard Distance tool in ArcGIS, respectively.
Development of simple sequence repeat markers
Simple sequence repeat (SSR) markers were developed using an improved technique for isolating codominant compound SSR markers (Lian and Hogetsu 2002; Lian et al. 2006). The total genomic DNA of V. grypoceras was extracted from leaves by using a modified cetyltrimethylammonium bromide (CTAB) method (Milligan 1992). An adaptor-ligated, restricted DNA library of V. grypoceras was then constructed according to the following procedure: DNA was separately digested with the blunt-end restriction enzymes, SspI, HaeIII, AfaI, and Alu. The restriction fragments were then ligated using a specific blunt adaptor (consisting of the 48-mer adapter strand 5′-GTAATACGACTCACTATAGGGCACGCGTGGTCGACGGCCCGGGCTGGT-3′ and an 8-mer adapter strand having the 3′-end capped with an amino residue, i.e., 5′-ACCAGCCC-NH2-3′) by using the DNA ligation kit (Takara). To block polymerase-catalyzed extension of the 8-mer adaptor strand, the ligated fragments were further treated with ddGTP by using AmpliTag Gold (Applied Biosystems). Fragments were amplified from the four digested DNA libraries by using compound SSR primer (AC)6(AG)5 or (TC)6(AC)5 and an adaptor primer (5′-CTATAGGGCACGCGTGGT-3′). The amplified fragments, ranging from 400 to 800 bp, were then separated on a 1.5 % LO3 agarose gel (Takara) and purified using the QIAquick Gel Extraction Kit (Qiagen). The purified DNA fragments were subsequently cloned using the QIAGEN PCR Cloning plus Kit (Qiagen) according to the manufacturer’s instructions. Briefly, polymerase chain reaction (PCR) products were ligated into the pDrive vector and transformed into QIAGEN EZ competent cells. Transformants were identified using blue/white screening on Luria–Bertani agar plates containing ampicillin, X-gal, and isopropyl-β-d-thiogalactopyranoside. The cloned fragments were amplified using the M13 forward and reverse primers from the plasmid DNA of positive clones. Amplified fragments were sequenced using the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems). For each fragment containing an (AC)6(AG)n or (TC)6(AC)n compound SSR sequence at one end, a specific primer was designed from the sequence flanking the compound SSR by using Primer3 (v. 0.4.0; National Human Genome Research Institute; Rozen & Skaletsky 2000). PCR amplifications were performed following the standard protocol of the Qiagen Multiplex PCR Kit (Qiagen) in a final volume of 6 µL, which contained 5 ng of extracted DNA, 3 µL of 2 × Multiplex PCR Master Mix, and 0.2 µM of each multiplexed primer. Compound SSR primers ((AC)6(AG)5 or (TC)6(AC)5) were labeled with fluorochromes 6-FAM or VIC (Applied Biosystems). PCRs were performed in a 5 µL volume containing 5 ng extracted DNA, 2 × Multiplex PCR Master Mix (2.5 µL), and 0.1 µM of each primer pair by using Qiagen Multiplex PCR kit (Qiagen). The reaction consisted of an initial denaturation step of 95 °C for 15 min; 25–26 cycles of denaturation at 94 °C for 30 s, annealing for 1.5 min, and extension at 72 °C for 1 min; and a final extension at 60 °C for 30 min. Annealing temperature (T a) and the number of cycles were optimized for each loci (Table 1). Amplified fragments were electrophoresed on an ABI PRISM 3130 Genetic Analyzer (Applied Biosystems). Fragment lengths were analyzed, and individuals were genotyped at six loci by using Gene Mapper version 4.0 (Applied Biosystems).
Genotyping
Genomic DNA was extracted using the CTAB protocol (Milligan 1992) and preserved with Tris–EDTA buffer (Tris–HCl 10 mM/EDTA 1 mM) at 4 °C. Six SSR markers developed in this study (Gry7, Gry18, Gry38, Gry95, Gry103, and Gry112; Table 1) were used to quantify the genetic diversity of all populations. We used the same genotyping procedures as used for the development of makers.
Assessment of genetic diversity
Three population indices of genetic diversity were calculated using MStool (Park 2001): average number of alleles per locus (A), observed heterozygosity (H O), and expected heterozygosity (H E). Allelic richness (A R) (El Mousadik and Petit 1996) and inbreeding coefficient (F IS; Wright 1969) were also computed using HP-Rare version 1.1 (Kalinowski 2004) and FSTAT version 2.9.3 (Goudet 1995). To test the independence of each loci, linkage disequilibrium between loci was tested with FSTAT (version 2.9.3; Goudet 1995) using data that excluded repeatedly observed genotypes.
Mann–Whitney U test was used to determine significant differences in A R and F IS between urban fragmented forests and suburban continuous forests by using R version 2.15.2 (R Development Core Team 2012).
Statistical model building
A statistical model was constructed in order to analyze the factors affecting genetic diversity of V. grypoceras in the urban forests. Genetic diversity was evaluated on the basis of the number of alleles as described below.
Probability of carrying a different allele can be regarded as independent. In the following equations, j represents number of alleles per locus. Y is defined as a variable of j. The probability distribution when Y = j can be described using the following binomial distribution:
where p is probability of carrying a different allele, and m is the number of leaf samples at each study site. When the number of samples is m, j can be an integer from 1 to 2 m (j = {1, 2,…, 2m}) because V. grypoceras is diploid and 2m is the maximum number of alleles per locus. In order to apply binomial distribution that assumes discrete numbers for k = {0, 1,…, n}, we defined k as 1 subtracted from j and n as 1 subtracted from 2m. Thus, X is defined as a variable of 1 subtracted from j.
The above description was used to construct a generalized linear mixed model (GLMM) with binomial distribution and logit link function to explain the genetic diversity of V. grypoceras in urban fragmented forests. We set the probability of carrying a different allele (p = k/n = (j − 1)/(2 m − 1); j, k, m, and n are as defined above) as the response variable. For explanatory variables of fixed effects, we adopted A SITE and D MT as spatial variables at the patch level; A VEG, A NOVEG, D O, |Z ANN|, D MAX, and D SD as spatial variables at the sub-patch level; and N P, N I, N IF, and N F as demographic variables. All the explanatory variables except |Z ANN| were log-transformed with a common logarithm before the analysis. Subsequently, we calculated the correlation coefficients for explanatory variables of fixed effects and selected sets of variables for model construction such that the correlation between the variables did not exceed ± 0.7. Finally, we standardized each explanatory variable of fixed effect in order to determine the most influential variables. For explanatory variables of random effects, we defined fragmented forests and loci (r PLACE and r LOCUS, respectively). We used the glmer function in the lme4 package, and the dredge function in the MuMIn package of R version 2.15.2 to construct statistical models of possible combinations of variables by avoiding the combinations of strong correlations (|r| > 0.7) to prevent problems with collinearity. The model with the lowest score of the Akaike’s information criterion (AIC) was retained as the final model (Akaike 1973). The final model was tested with the likelihood ratio test against the null model. Inclusion of random effects in the model was examined in preliminary tests and was found to improve the model. Explained deviance improved from 22.8 to 42.9 % by the incorporation of random effects.
Results
Field survey data
V. grypoceras was found in 16 of the 33 urban fragmented forests and 13 of the 14 suburban forests (Table 2). More than 30 individuals were observed in 12 fragmented forests and 12 suburban forests. The number of populations ranged from 2 to 35 and the area of vegetation from 1.6 to 47.2 ha in the 12 fragmented forests. Five fragmented forests (e, l, t, x, z) had a clustered distribution pattern of populations, two fragmented forests (g, v) had a dispersed pattern, and five fragmented forests (b, n, o, u, w) had a random pattern.
Development of SSR markers
We obtained six loci showing a clear, strong banding pattern for each allele (Table 1). The number of alleles per locus in all the studied populations ranged from 7 to 37 (Table 1). These polymorphisms of the developed markers were useful for evaluating the genetic diversity of V. grypoceras. The average number of alleles per locus at the population level ranged from 1.00 to 4.64, with the observed heterozygosity (H O) and expected heterozygosity (H E) at the population level ranging from 0.000 to 0.172 and from 0.000 to 0.762, respectively (Table 2). The F IS ranged from 0.27 to 1.00. The F IS values in most populations were high due to autogamy in cleistogamy flowers, which is well known in the genus Viola (Culley and Wolfe 2001). There was no evidence of significant linkage disequilibrium between any two loci, indicating the independence of each locus.
Comparison of genetic diversity indices between fragmented and suburban forests
There was no significant difference in A R (U = 54.5, p = 0.32 in the Mann–Whitney U test) and F IS (U = 69.5, p = 0.89) between the fragmented forests and suburban forests (Table 2; Fig. 3). The H O was low and F IS was high in most fragmented forests and suburban forests (Table 2).
Allelic richness (A R) and inbreeding coefficient (F IS) of Viola grypoceras var. grypoceras in the 12 urban fragmented forests and 12 suburban forests. The line inside the box indicates the median value. The bottom and top edges of the box indicate the 25th and 75th percentiles. The lower and upper limits of the whisker indicate the minimum and maximum, respectively. The circles indicate outliers
Factors affecting genetic diversity in fragmented forests
Correlation coefficients between explanatory variables of the fixed effects were calculated (Supplementary Material 1). Absolute values of correlation coefficients that were greater than 0.7 were the following combinations: A SITE–A VEG–A NOVEG, A VEG–D MAX–D SD, A NOVEG–|Z ANN|, and N P–N I–N IF–N F. Then, we constructed statistical models by avoiding the combinations of strong correlations (|r| > 0.7) and selected the final model with the lowest AIC. The final model that explained the genetic diversity in urban fragmented forests was constructed by selecting the following explanatory variables: A VEG, D O, |Z ANN|, r PLACE, and r LOCUS. The demographic variables such as N P, N I, N IF, and N F were not selected. The model was statistically significant against the null model as assessed by the likelihood test (p = 8.19 × 10−10). The residual deviance of this model was 60.2, and null deviance was 105.4: this model explained 42.9 % of all deviances. The analysis with standardization revealed that the order of influential variables was the following: A VEG, |Z ANN|, and D O (Table 3). The genetic diversity of V. grypoceras in urban fragmented forests tended to be higher if (1) the area of vegetation was larger; (2) the distribution pattern of populations in a forest was closer to random (i.e., intermediate distribution pattern between dispersed and clustered); and (3) the average distance between the nearest neighbor populations in a forest was shorter.
Discussion
Comparison of genetic diversity indices between fragmented and suburban forests
In general, genetic diversity is positively correlated with population size (Young et al. 1999). This means that fragmented small populations are likely to have low genetic diversity. In addition, rare alleles might be lost by genetic drift (Prober and Brown 1994). Thus, genetic diversity in fragmented forests is thought to be lower than that of un-fragmented forests. However, there was no significant difference of the genetic diversity in allelic richness between fragmented and suburban forests (Fig. 3).
Reproductive traits seem to provide a perspective about the susceptibility of a plant to fragmentation. In this study, the calculated F IS was suggestive of excessive inbreeding of V. grypoceras (Table 2). Given that this species produces cleistogamous flowers, inbreeding is likely to derive from self-pollination via cleistogamy, although the effects of low population size should be considered. This reproductive trait of V. grypoceras is considered to be relatively insusceptible to habitat fragmentation. This is supported by the finding of a meta-analysis on 52 plant species performed by Honnay and Jacquemyn (2007) that obligate or mainly out-crossing species are more vulnerable to the loss of genetic variation through habitat fragmentation than self-compatible species. Similarly, the genetic diversity of V. pubescence that has both chasmogamous and cleistogamous flowers did not appear to be impeded by fragmentation in the urban landscape of the USA (Culley et al. 2007), whereas Primula elatior, which is self-incompatible, showed decreased genetic diversity in small populations when recruitment occurred in forest fragments of Brussels urban zone, Belgium (Van Rossum 2008).
In addition, the trait that V. grypoceras forms a seed bank (Hosogi and Kameyama 2004) might also enhance population persistence. A persistent seed bank might mitigate the consequences of habitat fragmentation and protect a species from genetic drift and population genetic differentiation (Honnay et al. 2008).
Factors determining genetic diversity of V. grypoceras
Our results showed that the area of vegetation (A VEG) was the most influential variable to explain the genetic diversity of V. grypoceras in urban fragmented forests at Kyoto City, Japan (Table 3). Genetic diversity was higher when A VEG was larger. This suggests the possible deleterious effects of habitat fragmentation, more precisely, the effects of pure loss of habitat and reduced fragment size. The effect of increased spatial isolation of remnant fragments was not obvious because the distance from the nearest suburban mountain (D MT) was not selected as an explanatory variable in the result of GLMM.
We showed that the internal spatial arrangement of populations in a forest, i.e., the distribution pattern of populations and the distance between the nearest populations, affected the genetic diversity of V. grypoceras. Our results suggested that, when the distribution pattern of populations was closer to random (between dispersed and clustered patterns) and when the average distance between the nearest neighbor populations was shorter, genetic diversity tended to be higher.
Although the visual interpretation of the distribution of genotypes in each forest (Fig. 4) was supplemental and the effect of A VEG and other effects such as the genetic diversity of population founders and population history were included, we could identify several typical cases. In the dispersed pattern, genetic diversity within each population was low in the g forest. This might be because the long distance between populations makes frequent gene flow by cross-pollination difficult. In the clustered pattern, genotype within nearby populations was fixed in some cases (e, t, and z forests). Although the gene flow between nearby populations was easier than that in the dispersed populations, existing nearby populations might have resulted from the expansion of one genotype by self-pollination. On the other hand, in the random pattern, different alleles were found between intra- and inter-populations in some cases (n, o, and u forests). This could have been because gene flow between intra- and inter-populations might be afforded by moderate distances between the populations. Although the random distribution pattern of populations had higher genetic diversity, we found various cases in the actual genotype distribution (Fig. 4). The genetic diversity of founders of a population and population history are also thought to be important for forming the actual genotype distribution. For example, we could infer that the populations were likely to be formed from a single founder in t, K, L, and N forests since we detected only one genotype in each forest (Table 2).
Distribution of populations and genotypes of individuals sampled from urban fragmented forests. Alphabets filled in grey boxes represent forest IDs. Locations of populations were plotted on maps of fragmented forests. Different genotypes of sampled individuals were drawn using different symbols and linked to corresponding populations. Symbols are valid only within each forest and are not comparable between forests
Conclusions
We analyzed the factors affecting the genetic diversity of V. grypoceras in urban fragmented forests. We could not identify the effect of fragmentation by comparing the allelic richness between fragmented and suburban forests. This is probably because V. grypoceras has traits that allow it to tolerate fragmentation, such as traits of reproduction mainly by self-pollination via cleistogamy and of forming a seed bank. However, we could reveal that the area of vegetation, distribution pattern of populations, and average distance between the nearest populations affected the genetic diversity of this species in urban fragmented forests from the result of the GLMM. Therefore, we speculated that the deleterious effects of habitat fragmentation (pure loss of habitat and reduced fragment size) might exist in this species and might become more apparent if fragmentation further proceeds in the long term, even though allelic richness in urban forests was as high as that in the suburban forests at present. To our knowledge, this is the first report showing that the spatial distribution of populations within a forest affects the genetic diversity of a herbaceous species in urban fragmented forests. Considering the spatial arrangement of habitats in a forest will be required for better conservation planning of urban forests. For example, conservation of a single clustered area within a forest is likely to be insufficient; conservation of multiple habitats throughout a forest is more desirable. If the distance between populations is large, the preparation of additional habitats between populations is an option for genetic conservation.
References
Akaike H (1973) Information theory and an extension of the maximum likelihood principle: In Petrov BN, Caski F (eds) Proceedings of the 2nd international symposium information theory, Akadimiai Kiado, Budapest, pp 267–281
Alberti M (2005) The effects of urban patterns on ecosystem function. Int Region Sci Rev 28:168–192
Andren H (1994) Effects of habitat fragmentation on birds and mammals in landscapes with different proportions of suitable habitat: a review. Oikos 71:355–366
Berg G, Nordal I, Hestmark G (1998) The effect of breeding systems and pollination vectors on the genetic variation of small plant populations within an agricultural landscape. Oikos 81:17–29
Cruzan MB (2001) Population size and fragmentation thresholds for the maintenance of genetic diversity in the herbaceous endemic Scutellaria montana (Lamiaceae). Evolution 55:1569–1580
Culley TM, Wolfe AD (2001) Population genetic structure of the cleistogamous plant species Viola pubescens Aiton (Violaceae), as indicated by allozyme and ISSR molecular markers. Heredity 86:545–556
Culley TM, Sbita SJ, Wick A (2007) Population genetic effects of urban habitat fragmentation in the perennial herb Viola pubescens (Violaceae) using ISSR markers. Ann Bot 100:91–100
Culver DC, Beattie AJ (1978) Myrmecochory in Viola: dynamics of seed-ant interactions in some west Virginia species. J Ecol 66:53–72
Ebdon D (1985) Statistics in geography: Blackwell, Oxford, UK, pp 143–148
El Mousadik A, Petit RJ (1996) High level of genetic differentiation for allelic richness among populations of the argan tree Argania spinosa (L) Skeels endemic to Morocco. Theor Appl Genet 92:832–839
Fahrig L (2003) Effects of habitat fragmentation on biodiversity. Annu Rev Ecol Evol Syst 34:487–515
Fore SA, Hickey RJ, Vankat JL, Gunttman SI, Schaefer RL (1992) Genetic structure after forest fragmentation: a landscape ecology perspective on Acer saccharum. Can J Bot 70:1659–1668
Goddard MA, Dougill AJ, Benton TG (2010) Scaling up from gardens: biodiversity conservation in urban environments. Trends Ecol Evol 25:90–98
Godefroid S, Koedam N (2003) How important are large vs. small forest remnants for the conservation of the woodland flora in an urban context? Glob Ecol Biogeogr 12:287–298
Goudet J (1995) FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Hama E (2002) The wild violets of Japan in color: Seibundo Shinkosha, Tokyo (in Japanese), pp 2–11, 50–53
Hobbs RJ, Yates CJ (2003) Turner Review No. 7. Impacts of ecosystem fragmentation on plant populations: generalising the idiosyncratic. Aust J Bot 51:471–488
Honnay O, Jacquemyn H (2007) Susceptibility of common and rare plant species to the genetic consequences of habitat fragmentation. Conserv Biol 21(3):823–831
Honnay O, Hermy M, Coppin AP (1999) Effects of area, age and diversity of forest patches in Belgium on plant species richness, and implications for conservation and reforestation. Biol Conserv 87:73–84
Honnay O, Bossuyt B, Jacquemyn H, Shimono A, Uchiyama K (2008) Can a seed bank maintain the genetic variation in the above ground plant population? Oikos 117:1–5
Horning ME, Webster MS (2009) Conservation genetics of remnant Lilium philadelphicum populations in the Midwestern United States. Am Midl Nat 161:286–300
Hosogi D, Kameyama A (2004) Timing for the collection of topsoil from a deciduous forest for use as planting material in suburban Tokyo, Japan. Ecol Eng 23:371–386
Imanishi A, Imanishi J, Murakami K, Morimoto Y (2005) A study for conservation of herbaceous plants at urban green spaces: a comparison to suburban secondary forest. J Landsc Archit Asia 1:127–132
Kalinowski ST (2004) Counting alleles with rarefaction: private alleles and hierarchical sampling designs. Conserv Genet 5:539–543
Kwak MM, Velterop O, van Angel J (1998) Pollen and gene flow in fragmented habitats. Appl Veg Sci 1:37–54
Ledig FT (1992) Human impacts on genetic diversity in forest ecosystems. Oikos 63:87–108
Lian C, Hogetsu T (2002) Development of microsatellite markers in black locust (Robinia pseudoacasia) using a dual-suppression-PCR technique. Mol Ecol Notes 2:211–213
Lian C, Wadud MA, Geng Q, Shimatani K, Hougetsu T (2006) An improved technique for isolating codominant compound microsatellite markers. J Plant Res 119:415–417
McKinney ML (2002) Urbanization, biodiversity, and conservation. Bioscience 52:883–890
Milligan B (1992) Plant DNA isolation. In: Hoelzel AR (ed) Molecular genetic analysis of populations: a practical approach. IRL Press, Oxford, pp 59–88
Mills LS, Tallmon DA (1999) The role of genetics in understanding forest fragmentation. In: Rochelle JA, Lehmann LA, Wisniewski J (eds) Forest fragmentation: wildlife and management implications. Brill, Boston, pp 171–186
Oostermeijer JGB, Luijten SH, den Nijs JCM (2003) Integrating demographic and genetic approaches in plant conservation. Biol Conserv 113:389–398
Park SDE (2001) Trypanotolerance in West African cattle and the population genetic effects of selection. Dissertation, University of Dublin
Prober SM, Brown AHD (1994) Conservation of the grassy white box woodlands: populations genetics and fragmentation of Eucalyptus albens. Conserv Biol 8:1003–1013
R Development Core Team (2012) R-2.15.2: A language and environment for statistical computing. R foundation for Statistical Computing, Vienna, Austria
Van Rossum F (2008) Conservation of long-lived perennial forest herbs in an urban context: Primula elatior as study case. Conserv Genet 9:119–128
Whigham DF (2004) Ecology of woodland herbs in temperate deciduous forests. Annu Rev Ecol Evol Syst 35:583–621
Wright S (1969) Evolution and the genetics of population. The theory of gene frequencies, vol 2. University of Chicago Press, Chicago
Young AG, Merriam HG, Warwick SI (1993) The effects of forest fragmentation on genetic variation in Acer saccharum Marsh. (sugar maple) populations. Heredity 71:277–289
Young A, Boyle T, Brown T (1996) The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol 11:413–418
Young AG, Brown AHD, Zich FA (1999) Genetic structure of fragmented populations of the endangered daisy Rutidosis leptorrhynchoides. Conserv Biol 13:256–265
Acknowledgments
This work was supported by JSPS KAKENHI grant number 22241010.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Toma, Y., Imanishi, J., Yokogawa, M. et al. Factors affecting the genetic diversity of a perennial herb Viola grypoceras A. Gray var. grypoceras in urban fragmented forests. Landscape Ecol 30, 1435–1447 (2015). https://doi.org/10.1007/s10980-015-0197-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10980-015-0197-1