Abstract
Trehalose is a unique disaccharide capable of protecting proteins against environmental stress. A novel trehalose synthase (TreS) gene from Rhodococcus opacus was cloned and expressed in Escherichia coli Top10 and BL21 (DE3) pLysS, respectively. The recombinant TreS showed a molecular mass of 79 kDa. Thin layer chromatography (TLC) result suggested that this enzyme had the ability to catalyze the mutual conversion of maltose and trehalose. Moreover, high-performance liquid chromatography (HPLC) result suggested that glucose appeared as a byproduct with a conversion rate of 12 %. The purified recombinant enzyme had an optimum temperature of 25 °C and pH optimum around 7.0. Kinetic analysis revealed that the K m for trehalose was around 98 mM, which was a little higher than that of maltose. The preferred substrate of TreS was maltose according to the analysis of k cat/K m. Both 1 and 10 mM of Hg2+, Cu2+ and Al3+ could inhibit the TreS activity, while only 1 mM of Ca2+ and Mn2+ could increase its activity. Five amino acid residues, Asp244, Glu286, Asp354, His147 and His353, were shown to be conserved in R. opacus TreS, which were also important for α-amylase family enzyme catalysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Trehalose composed of two glucose molecules is a non-reducing disaccharide, which has been isolated from many organisms ranging from bacteria, yeast, fungi to higher and lower of animals and plants [7]. Trehalose and its derivatives can play a role as protectants against various physical and chemical stresses, such as cold, heat, dehydration and oxygen radical [22]. Moreover, trehalose works as an important cell constituent of many Mycobacteria and Corynebacteria strains [2]. Due to its properties, it can be used as a useful supplement in industrial applications, like foods, cosmetics and pharmaceutical industries [23].
Five distinct biosynthetic pathways can lead to the formation of trehalose: (a) TPS-TPP [3]; (b) TreP [25]; (c) TreT [28]; (d) TreY-TreZ [8]; and (e) TreS [30]. TreS is an enzyme that allows one-step formation of trehalose from maltose through intramolecular transglycosylation. In summary, this pathway is capable of being used in the industrial manufacture of trehalose [29, 31]. Members of the Rhodococcus genus have a marked ability to metabolize a wide variety of xenobiotic compounds, and can survive in extreme environment, such as R.opacus PD630 could withstand water stress [1]. The formation of biosurfactants by rhodococci has been studied well, and several trehalose-containing glycolipids that often occur as a complex mixture during growth of rhodococci were structurally clarified. Certain trehalose lipids from rhodococci also played an important role in many asides [10, 21].
Rhodococcus opacus B4 was isolated in 2005, which had a tolerance to organic solvents like benzene, toluene, ethyl-benzene, xylene and styrene and could survive in the extreme environment for at least 5 days [19, 20, 26]. This tolerance probably attributed to glycolipids, typically trehalose mycolates, which were the parts of its own cell envelope [27].
The genome sequence of R. opacus B4 (EMBL AP011115) was published 3 years ago. Nevertheless, very little work has been presented on the biochemical and biophysical properties of its putative TreS. In our study, a novel TreS gene was cloned from the genomic DNA of R. opacus ACCC 41021and overexpressed under the control of T7 promoter in Escherichia coli. We studied the biochemical function and enzyme features of this TreS. To our knowledge, this is the first report on TreS from Rhodococcus genus and would provide helpful information for the studies both on TreS and on Rhodococcus in the future.
2 Materials and Methods
2.1 Bacterial Strains, Plasmids and Cultivation
Rhodococcus opacus was obtained from the American Type Culture Collection (ACCC 41021). The pGEM Easy-T Vector System (Promega A3600, USA) was used as a cloning vector. The pRSET-B(Novagen, USA)plasmid was used as an expression vector, which contained a T7 promoter and a hexahistidine-tag. The bacterial strains E. coli Top10 and BL21 (DE3) pLysS (Novagen, USA) were used as hosts for target gene clone and expression, respectively. The E. coli strains were grown either in Luria–Bertani (LB) broth or on LB agar supplemented with 100 μg/mL ampicillin (Amp) at 37 °C. The R. opacus strain was cultured at 26 °C in ISP-1 medium (glucose 0.4 %, yeast extract 0.4 %, malt extract 1 % and CaCO3 0.2 %).
2.2 Construction of Recombinant Expression Vector
The genomic DNA of R. opacus ATCC 41021 was obtained by a bacteria genome DNA extracting kit (Tiangen Biotech, China) and worked as the template for amplification of target gene. The oligonucleotide primers for amplification were designed as 5′-GAAGATCTATGCCCCTGGAAGATTCCTCGTC-3′ (BglII cleavage site underlined) and 5′-CCAAGCTTTCACTGTTGCACCGCCTGTT-3′ (HindIII cleavage site underlined). The amplified DNA fragments were linked with pGEM Easy-T vector and transformed into E. coli Top10. The correct transformants were digested with BglII and HindIII, then being linked with pRSET-B that had been digested with the corresponding restriction enzymes. The correct recombined vector was transformed into E. coil BL21 (DE3) pLysS to express recombinant TreS.
2.3 Expression of the Recombinant TreS
The recombinant E. coil BL21 (DE3) pLysS containing the TreS gene was cultured in 1 L LB broth supplemented with 100 μg/mL Amp and grew until reaching OD600 of 0.6. Then, isopropyl β-D-1-thiogalactopyranoside (IPTG, final concentration 0.5 mM) was added to induce the protein expression, and incubation was continued for 6 h at 25 °C. The cells were harvested by centrifugation at 4,000 rpm for 10 min at 4 °C and the resulting cell pellet was stored at −75 °C.
2.4 Protein Purification
Each pellet was resuspended in 20 mL lysis buffer (50 mM K2HPO4-KH2PO4, pH 8.0, 0.5 M NaCl and 10 % glycerol). Phenylmethanesulfonyl fluoride (PMSF, final concentration 1 mM) and lysozyme (0.4 mg/mL) were supplemented. After half-hour incubation on ice, cells were lysed using a sonicator (Scientz JY92-DN, China). After that, 10 % Triton X-100 (0.05 %, final concentration), MgCl2 (final concentration 1 M), and DNase (10 μg/mL) were supplemented. Cells were lysed after incubation again on ice. The lysates were harvested by centrifugation (6,000 rpm, 4 °C and 20 min), and then applied to NTA nickel-ion column (Novagen, USA), which had been washed twice with 10 bed volumes of lysis buffer. The column containing TreS was washed by lysis buffers containing different concentrations (20–500 mM) of imidazolea at the flow speed of 0.2 mL/min. Finally, the solution containing target protein was concentrated by ultrafiltration (MilliporeUFC803024, BJXJKSW, and China) following the manufacturer’s instructions.
2.5 Electrophoretic Methods: SDS-PAGE, Native-PAGE of the Recombinant TreS
Protein samples from expression and purification (10 μL) were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on polycarylamide gel (12 %) in accordance with Laemmli [12]. Electrophoresis was performed on 1.5 mm vertical gel slabs at 24 mA for 4 h. Gels were stained with Coomassie Brilliant blue G250 after electrophoresis. The standard protein mixtures obtained from TransGen Biotech (Beijing, China) were used as reference.
The oligomerization degree of the recombinant TreS was determined by native gel electrophoresis employing a 12 % gel according to native Laemmli system [12] by using native protein marker (TIANGEN Biotech CO, China). The Bradford method [4] was used to quantify the recombinant protein with bovine serum albumin as a standard.
2.6 Activity Assay of TreS
The enzymatic activity of TreS was identified by the method of thin layer chromatography (TLC). The reaction mixtures containing either 90 mM maltose or trehalose and 100 μL enzyme solution were performed at 25 °C for 1 h. To stop the enzymatic reaction, the reaction mixtures were heated at 100 °C for 10 min. The products (1 μL) were injected onto silica gel plates (HPTLC 10 × 20 cm, Merck, Germany) and a hair drier was used to dry the plates. To separate the samples, a solvent system of 1-butanol/pyridine/water (4/5/1) was used. The plates were dried again by a hair drier after 1 h. Sugars were detected by spraying the plate with 20 % (v/v) sulfuric acid in methanol. To view the results, the plate was heated in 105 °C for 5–10 min. The BandScan software was used to quantify the reaction mixtures. One unit of enzyme activity was defined as the amount of enzyme that generated 1 mmol trehalose per minute. HPLC system equipped with a Dionex 2500 system was used to confirm and quantify the products of different reactions. Ten pmol of reaction samples were injected into the system at a flow speed of 10 μL per minute.
The effects of metal ions on the activity of TreS were determined in an assay buffer containing 1 or 10 mmol/L metal ions under the standard reaction conditions.
2.7 Effects of Temperature on TreS
A reaction mixture containing 90 mM maltose was incubated with 100 μL enzyme solution at different temperatures (15–70 °C) to identify the effects of temperature on TreS activity. To study the stability of TreS, reaction mixtures were pre-incubated at different temperatures (15–70 °C) for 1 h, and then examined the residual activities under the standard assay. All reactions were carried out at pH 7.0. To stop the enzymatic reaction, the reaction mixtures were heated in 100 °C for 10 min. The mixtures were analyzed by TLC and HPLC.
2.8 Effects of pH on TreS
A reaction mixture (pH 3–11) containing 90 mM maltose was incubated with 100 μL enzyme solution at 25 °C for 1 h to identify the effects of pH on TreS activity. To study the stability of TreS, the reaction mixtures were pre-incubated at different pHs (pH 3–11) for 1 h, and then the residual activities were examined under the standard assay. To stop the enzymatic reaction, the reaction mixture was heated in 100 °C for 10 min.
2.9 Determination of Kinetic Parameters
The K m, V max, and k cat of the TreS were analyzed according to the method of Lineweaver and Burk [15]. The analyses of Kinetic parameters were carried out under the conditions of pH 7.0 and 25 °C for 1 h in 100 mmol/L potassium phosphate buffer containing different substrates (maltose, trehalose, and glucose) at different concentrations (1–50 mM).
3 Results
3.1 Cloning of TreS Gene From R. opacus
Since trehalose has been detected in R. opacus [1], the biosynthesis of trehalose was assumed in this organism. Besides, R. opacus B4 was reported to have the TreS gene according to its genomic DNA sequence (EMBL AP011115). Hence, we designed primers according to the TreS gene sequence of R. opacus B4. As a result, we successfully isolated the TreS gene from R. opacus ACCC 41021, which had a length of 1,857 bp and encoded 618 amino acids. The TreS gene had been submitted to the GenBank database, and the Accession Number was KC473564. This TreS showed 96, 93, and 81 % homology with the TreS genes from Rhodococcus jostii RHA1 (gi = 110816552), R. opacus B4 (gi = 226359415), and Pseudonocardia dioxanivorans CB1190 (gi = 326948588), respectively. On the other hand, this gene produce showed 99, 98, 96, and 79 % identify with R. opacus M213 (gi = 419961084), Rhodococcus jostii RHA1 (gi = 111023004), Rhodococcus opacuas B4 (gi = 226365509), and Pseudonocardia dioxanivorans CB1190 (gi = 331695578), respectively. The product of TreS gene had no signal peptide according to the analysis of its amino acid sequence, which suggested that this amino acid sequence was an intracellular protein.
3.2 Expression and Purification of the TreS
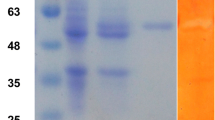
With the help of T7 promoter, the recombinant strain transformed with plasmid pRSET-B that carried the TreS gene had successfully expressed our aim gene and a hexabistidine-tagged. The enzyme was purified by NTA nickel-chelat chromatography, and most of TreS was eluted with lysis buffer consisting of 100 mM imidazole, 50 mM K2HPO4-KH2PO4, 0.5 M NaCl and 10 % glycerol. This recombinant protein showed a 76 kDa band on SDS-PAGE gel (Fig. 1), which was a little larger than 71 kDa predicted by Compute pI/Mw (http://web.expasy.org/compute_pi/). This phenomenon was due to the extra parts coming from the pRSET- B vector. However, in native condition, TreS was present in a tetramer oligomeric state with a molecular weight of 290–300 kDa (date not shown), which kept pace with the TreS gene coming from Meiothermus ruber [31]. The fusion protein reached its peak expression level at 25 °C and induced for 6 h with 0.5 mM IPTG. The fusion protein accounted for approximately 45 % of total protein content in the bacterial lysates (analyzed by BandScan). The cellular extract of E.coli harboring TreS had a specific activity of 3.2 U/mg, which was greater than 2.822 and 0.726 U/mg coming from S. solfataricus KM1 and MT4 enzymes [24].
SDS-PAGE analysis for the recombinant protein. Lane 1, 2: Total cell proteins of control group in supernatant and deposit, inducing for 4 h by IPTG; Lane 3, 4: Total cell proteins of positive group in supernatant and deposit, after inducing for 0 h by IPTG; Lane 5, 6: Total cell proteins of positive group in supernatant and deposit, after inducing for 6 h by IPTG; Lane 7: Protein molecular weight marker (80, 60, 40, 30, 20 kDa); Lane 8 The purified protein
3.3 Activity of the Recombinant Enzyme
The products of reactions were showed in Fig. 2. TLC results revealed that this recombinant enzyme had the ability to catalyze both maltose and trehalose, moreover, these phenomenon were also verified by the analysis of HPLC. To quantify the trehalose production, the reaction mixtures were analyzed by HPLC. HPLC results showed additional that glucose appeared as a byproduct, which was in line with the TreS coming from Arthrobacter aurescens [29]. The HPLC results also showed the transformation efficiency of trehalose and glucose was 67 and 12 %, respectively.
TLC analysis of reaction products from different substrates by TreS. Lane 1: standard molecule of maltose; Lane 2: standard molecule of trehalose; Lane 3: maltose as the substrate; Lane 4: trehalose as the substrate. The reaction mixtures containing 90 mM substrate and 100 μL enzyme solutions were performed at 25 °C for 1 h
3.4 Effects of Temperature on TreS
The effects of temperature on TreS were showed in Fig. 3. The optimal temperature (25 °C) of recombinant enzyme was lower than many other TreS such as the one (50 °C) coming from Meiothermus ruber [31] and the other (35 °C) coming from Corynebacterium glutamicum [9]. Upon prolonged incubation of TreS at the temperature range of 15–70 °C, the stability of TreS remained constant at the temperature range of 15–45 °C, while the enzymatic activity of TreS descended dramatically in 3 h when the temperature was above 60 °C. TreS catalyzed both transglycosylation reaction to produce trehalose and hydrolysis reaction to split maltose to glucose [9]. The trehalose yield increased along with a lower temperature, due to the hydrolysis reaction decreased [6]. The structural flexibility of TreS decreased at a lower temperature according to Koh et al. [11], thus, the reaction temperature should be reduced to get the best conversion yield.
Effects of temperature on TreS with maltose as the substrate. The effects of various temperatures (15–70 °C) on the activity of TreS were incubated at pH 7.0 for 1 h, using maltose as the reaction substrate. To study the stability of TreS, reaction mixtures were pre-incubated at different temperatures (15–70 °C) for 1 h, and then examined the residual activities
3.5 Effects of pH on TreS
The optimal pH was obtained by the analysis of activity at the pH range of 3–11 (Fig. 4). The pH dependence of the TreS activity is a typical bell-shaped curve suggesting the enzymatic hydrolysis depends upon two ionizable amino acid residues. And the pH optimum of this enzyme was around pH 7.0. Upon prolonged incubation of TreS in the pH range of 4.0–6.5 and 10.5–11.0 the enzyme lost 30–60 % of its activity in 1 h, whereas in the pH range of 6.5–8.5, the enzyme activity remains constant. The enzyme maintained the highest stability at pH 7.0–7.5, which was consistent with the properties of other TreS [9].
Effects of pH on TreS with maltose as substrate. The effects of different pHs (pH 3–11) on the activity of TreS were studied at 25 °C for 1 h, using maltose as the reaction substrate. To study the stability of TreS, the reaction mixtures were pre-incubated at different pHs (pH 3–11) for 1 h, and then examined the residual activities
3.6 Effects of Metal Ions and Reagents
It’s well known that an enzyme activity was frequently affected by metal ions and reagents [5]. To detect the influences of metal ions and reagents on TreS activity, we tested enzyme activity under the conditions containing different metal ions and reagents (1 or 10 mM), shown in Fig. 5. TreS activity could been suppressed by 1 mM Hg2+, Cu2+, SDS and Al3+, while increased by 1 mM Ca2+ and Mn2+. But almost all metals and reagents inhibited the enzyme activity when the concentration reached 10 mM, except for the chelator EDTA, which suggested that the enzyme was not sensitive to EDTA.
3.7 Kinetic Parameters
Reactions of the recombinant TreS with different substrates were performed (Table 1). The enzyme TreS had a higher affinity for maltose than trehalose according to their Km values. It had 4.3-fold higher enzyme efficiency toward maltose than trehalose according to k cat/K m, which indicated that the reaction equilibrium trend was producing trehalose. With glucose adding in, the Km presented a five-fold increase, while k cat/K m decreased by six-fold, implying that glucose could inhibit the formation of trehalose.
4 Discussion
Two TreS genes, 1,860 bp gene (gi = 226359415) and 2,190 bp gene (gi = 226359415), were previously known about the genomic DNA of R. opacus B4 (EMBL AP011115). In our study, we successfully cloned the 1,857 bp gene from R. opacus ACCC 41021 according to the 1,860 bp gene, which encoded 618 amino acids. It was more similar to other known TreS genes than the 2,190 bp gene [16, 31].To our known, members of Rhodococcus had the ability to generate biosurfactant molecules [13]. The molecules were mainly glycolipids, especially trehalose monomycolates, trehalose dimycolates, trehalose trimycolates, as well as octa-acylated derivatives of trehalose [14, 18, 21]. Furthermore, the extracs of rhodococcal cell envelope were normally represented by trehalose mycolates [27]. According to this phenomenon, R. opacus was a good candidate to isolate the gene encoding an enzyme for synthesizing trehalose.
Besides, this TreS belonged to the amylase family according to the analysis of N-terminal by BLAST [17]. Based on the result of Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/), we found five highly conserved regions, which were identical to the α-amylase family (Table 2) [17]. The important residues are Asp244, Glu286, Asp354, His147 and His353. Asp244 and Asp354 might function as nucleophiles that attack the bonded anomeric carbon of maltose. Glu286 might play a role in proton donor. His147 and His353 might act as the substrate binding site [6]. The role of conserved sites also confirmed in Thermus caldophilu [11] and Meiothermus ruber [31]. We tested both the effects of temperature and pH on TreS activities and kinetics parameters of TreS to get the properties of this enzyme. Concluding the data from the pH/temperature dependence and stability studies, the optimum temperature of TreS was 25 °C and the optimal pH of TreS was 7.0. Besides, the TreS was stable with the highest activity at 15–30 °C and pH 6.5–8.5 for several hours, which was related to the environmental conditions of this species [24]. It could produce 67 % trehalose and 12 % glucose using maltose as substrate. Lower reaction temperature could decrease the weak hydrolytic property and decrease the synthesis of glucose [11]. Considering the properties of this protein, the R. opacus TreS has the potential to be used in industrial applications.
Abbreviations
- Amp:
-
Ampicillin
- ATCC:
-
American type culture collection
- EMBL:
-
The European Molecular Biology Laboratory
- HPLC:
-
High-performance liquid chromatography
- IPTG:
-
Isopropyl β-D-1-thiogalactopyranoside
- LB:
-
Luria–Bertani
- PCR:
-
Polymerase chain reaction
- SDS-PAGE:
-
SDS polyacrylamide gel electrophoresis
- TLC:
-
Thin layer chromatography
- TreS:
-
Trehalose synthase
References
Alvarez HM, Silva RA, Cesari AC, Zamit AL, Peressutti SR, Reichelt R, Keller U, Malkus U, Rasch C, Maskow T, Mayer F, Steinbuchel A (2004) FEMS Microbiol Ecol 50:75–86
Aranda FJ, Teruel JA, Espuny MJ (2007) Chem Phys Lipids 149:S23–S24
Avonce N, Mendoza-Vargas A, Morett E, Iturriaga G (2006) BMC Evol Biol 6:109
Bradford MM (1976) Anal Biochem 72:248–254
Buisson G, Duee E, Haser R, Payan F (1987) EMBO J 6:3909–3916
Chen YS, Lee GC, Shaw JF (2006) J Agric Food Chem 54(19):7098–7104
Elbein AD, Pan YT, Pastuszak I, Carroll D (2003) Glycobiology 13:17R–27R
Fang TY, Hung XG, Shih TY, Tseng WC (2004) Extremophiles 8:335–343
Kim TK, Jang JH, Cho HY, Lee HS, Kim YW (2010) Food Sci Biotechnol 19:565–569
Kitamoto D, Isoda H, Nakahara T (2002) J Biosci Bioeng 94:187–201
Koh S, Kim J, Shin HJ, Lee D, Bae J, Kim D, Lee DS (2003) Carbohyd Res 338:1339–1343
Laemmli VK (1970) Nature 227:680–685
Lang S, Philp JC (1998) Anton Leeuw Int J G 74:59–70
Larkin MJ, Kulakov LA, Allen CCR (2006) Adv Appl Microbiol 59:1–29
Lineweaver H, Burk D (1934) J Am Chem Soc 56:658–666
Ma Y, Xue L, Sun DW (2006) J Food Eng 77:342–347
MacGregor EA, Janecek S, Svensson B (2001) Biochim Biophys Acta 1546:1–20
Martinkova L, Bronislava U, Miroslav P, Nesvera J, Kren V (2009) Environ Int 35:162–177
Na KS, Kuroda A, Takiguchi N, Ikeda T, Ohtake H, Kato J (2005) J Biosci Bioeng 99:378–382
Na KS, Nagayasu I, Kuroda A (2005) J Biosci Bioeng 99:408–414
Niescher S, Wray V, Lang S, Kaschabek SR, Schlomann M (2006) Appl Microbiol Biotechnol 70:605–611
Purvis JE, Yomano LP, Ingram LO (2005) Appl Environ Microb 71:3761–3769
Richards AB, Krakowka S, Dexter LB, Schmid H, Wolterbeek APM, Waalkens-Berendsen DH, Shigoyuki A, Kurimoto M (2002) Food Chem Toxicol 40:871–898
Schiraldi C, Di LI, De RM (2002) Trends Biotechnol 20:420–425
Schwarz A, Goedl C, Minani A, Nidetzky B (2007) J Biotechnol 129:140–150
Shiho Y, Masafumi S, Yoshihiro I, Kohsuke H, Yuka S, Takeshi O, Junichi K, Hisao O (2007) Appl Microbiol Biotechnol 74:761–767
Sutcliffe IC (1998) Antonie Van Leeuwenhoek 74:49–58
Tzvetkov M, Klopprogge C, Zelder O, Liebl W (2003) Microbiol-SGM 149:1659–1673
Wu XL, Ding HB, Yue M, Qiao Y (2009) Appl Microbiol Biotechnol 83:477–482
Zdzieblo A, Synowiecki J (2006) Food Chem 96:8–13
Zhu YM, Wei DS, Zhang J (2010) Extremophiles 14:1–8
Acknowledgments
This research was supported by Hi-Tech Research and Development Program of China (Grant No. 2012AA021502; 2011BAD26B002), and the natural science foundation of Jiangsu Province (BK2011099) and the basic research grant for central non-commercial research institutes (2012ZL099).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yan, J., Qiao, Y., Hu, J. et al. Cloning, Expression and Characterization of a Trehalose Synthase Gene From Rhodococcus opacus . Protein J 32, 223–229 (2013). https://doi.org/10.1007/s10930-013-9476-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-013-9476-3