Abstract
This paper presents a novel algorithm (CVSTSCSP) for determining discriminative features from an optimal combination of temporal, spectral and spatial information for motor imagery brain computer interfaces. The proposed method involves four phases. In the first phase, EEG signal is segmented into overlapping time segments and bandpass filtered through frequency filter bank of variable size subbands. In the next phase, features are extracted from the segmented and filtered data using stationary common spatial pattern technique (SCSP) that can handle the non- stationarity and artifacts of EEG signal. The univariate feature selection method is used to obtain a relevant subset of features in the third phase. In the final phase, the classifier is used to build adecision model. In this paper, four univariate feature selection methods such as Euclidean distance, correlation, mutual information and Fisher discriminant ratio and two well-known classifiers (LDA and SVM) are investigated. The proposed method has been validated using the publicly available BCI competition IV dataset Ia and BCI Competition III dataset IVa. Experimental results demonstrate that the proposed method significantly outperforms the existing methods in terms of classification error. A reduction of 76.98%, 75.65%, 73.90% and 72.21% in classification error over both datasets and both classifiers can be observed using the proposed CVSTSCSP method in comparison to CSP, SBCSP, FBCSP and CVSCSP respectively.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
For people with neurological disorders, brain computer interface (BCI) has provided a potential way to establish communication and restore lost motor functions by translating their brain signals into device commands. BCI has garnered much interest among researchers due to its practical applications in computers, virtual gaming, assistive appliances, speech synthesizers, and neural prostheses [1,2,3,4,5]. Several brain modalities have been used to measure brain signals such as magnetoencephalography (MEG), functional magnetic resonance imaging (fMRI), positron emission tomography (PET), electroencephalography (EEG), etc. Among these modalities, EEG based BCI is the most widely used modality for analysis of brain signals due to its non-invasive nature, low measurement cost and high resolution. In EEG based BCI, electrodes are placed on the scalp of the brain to capture electrical signals, that are generated by the neuronal activity of brain, for the purpose of communication. Major EEG based BCI paradigms include P300, visually evoked potential, sensorimotor rhythms (motor–imagery) [6], etc. Among these, particular attention has been received by motor-imagery based BCIs, which involve visualization of the movement of a specific body motor part [7, 8]. Motor imagery BCIs use variations in sensory motor rhythms (μ and β rhythms) to translate brain signals into control commands [9]. These variations are detected over the sensorimotor cortex and are induced by execution or imagination of hand or leg movement [6]. The amplitude of sensorymotor rhythms reduces during motor imagination or execution which is known as Event-Related Desynchronization (ERD). The subsequent increment in the amplitude of sensorymotor rhythms instantly after the execution or imagination of movement is called as Event-Related Synchronization (ERS) [6, 10,11,12].
These brain signals undergo volume conduction which provides weak spatial resolution [6, 11]. To analyze a single trial EEG, a BCI device is tuned to the subject specific characteristics by calculating data dependent spatial filters. CSP method [2, 13, 14], a data-driven spatial filtering technique, is a widely used spatial filter method in motor imagery BCI. CSP is aimed at finding the spatial filters from multichannel EEG signal which maximize variance of one class and at the same time minimizes variance of the other class [2, 6, 14]. The method is computationally simple and reflects the specific activation of cortical areas by assigning weights to the electrodes according to their importance. It also reduces the dimension of the data.
The major drawback of the CSP method is that it is sensitive to the presence of artifacts in raw EEG data and the non-stationary nature of EEG signals. Further, CSP suffers from the small size problem as the number of task-related trials is less as compared to the number of electrodes [15]. The size of covariance matrices obtained using CSP is of the order O(N2), where N is the number of electrodes. If the covariance matrices in CSP are estimated with a relatively small number of trials, the presence of a single trial contaminated with artifacts may not provide an appropriate CSP filter. In such situation, CSP suffers from the problem of overfitting [16, 17] and thus leads to poor performance. A variant of CSP, Stationary CSP (SCSP), is suggested in literature [17] that can handle the artifacts and non-stationarity of EEG data.
However, CSP as well as SCSP, in its original formulation, do not consider the spectral information of the signal to derive the spatial filter. It is pointed out in the literature [13, 14] that a specific set of frequency bands helps in discriminating the two different motor imagery tasks. A set of the frequency range for EEG is defined according to its distribution over the scalp or biological significance [6, 18]. These frequency bands are referred as delta (1–4 Hz), theta (4–7 Hz), alpha (7–12 Hz), beta (12–30 Hz), and gamma (30–40 Hz). Most of the relevant information gained from motor imagery signals lies in mu (7–12 Hz) and beta (12–30 Hz) bands of brain EEG which typically falls in the frequency bands of 7–30 Hz [2]. CSP spatial filters are thus applied to EEG signals filtered from these relevant frequency bands (mu and beta bands) to optimize the performance of a motor imagery BCI. It is possible that frequency subbands other than mu (7–12 Hz) and beta (12–30 Hz) are more relevant to distinguish motor imagery tasks. Manual tweaking, as well as exhaustive search, can help in determining the best frequency bands, but this is computationally intensive. It is thus desirable to automatically find optimal subject-specific frequency bands that relate to brain activities associated with motor imagery tasks in order to achieve higher accuracy.
In literature, several research works [19,20,21] suggested methods to determine spatial filters from a predefined filter bank of fixed sized non-overlapping subband frequency filters. Working in this direction, Novi et al. (2007) proposed Subband CSP (SBCSP) [19] that used data filtered from different fixed sized subbands to extract features by applying CSP followed by linear discriminant analysis to distinguish motor imagery tasks. The features from subbands were ranked using recursive feature elimination (RFE) method based on SVM. In Filter Bank CSP (FBCSP) method [20], maximal mutual information criterion was used to select optimal spatio-temporal filters from data filtered using different fixed size frequency bands. SBCSP and FBCSP methods employ manual setting of fixed sized (bandwidth 4 Hz) frequency subbands in the range of 4–40 Hz. Thus, any other relevant subbands that can possibly be present in the given frequency band range were not explored in these two methods, which may lead to poor performance.
The efficacy of the CSP/SCSP also depends on the choice of time segment of the EEG taken relative to the visual cue presented to the subject [22, 23]. Typically, time segment of 1 s after the cue is taken for computation of CSP/SCSP spatial filters. However, the generation of motor imagery related EEG rhythms varies with the subject involved and relevant time segment cannot be identified manually [22]. Thus, there is a need to identify subject specific and task related frequency filter and relevant time segment EEG data for better performance of motor imager tasks.
To determine the subject specific optimal frequency bands, the research work [21] has proposed the Combined Variable Sized Common Spatial Patterns method (CVSCSP) that generates a variable size frequency subbands filter bank. However, CVSCSP is not able to detect the irregularities in performance of a given subject that arise due to use of irrelevant time segment of a trial. Further, it cannot handle the artifacts and non-stationarity of the signal. In this paper, we have proposed a modified version of the CVSCSP that is more robust to artifacts and include utilizes relevant temporal features. In the proposed method, in order to capture the relevant temporal features for a given subject, we segment the data from each trial into three different overlapping time segments. The obtained data from each time segment is then bandpass filtered using the variable size frequency subbands. Spatial features are extracted using SCSP to handle artifacts and nonstationarity from the bandpass filtered data of different time segment separately. Finally, the extracted features are combined to form a high dimensional feature vector. Thus, the proposed model is able to take an advantage of temporal, spatial and frequency information of the data simultaneously. However, the high dimension feature vector obtained may contain irrelevant features. In order to obtain relevant subset of features, univariate feature selection method is used to rank the obtained features. We have investigated four well-known univariate feature selection methods to rank features.
The proposed method involves four phases: In the first phase, we segment the raw data into overlapping time segment data, generate filter bank of variable sized frequency subbands to filter the data. In the second phase, a combination of SCSP and linear discriminant analysis is used to compute features from filtered data. The obtained features are ranked using univariate feature selection method in the third phase. Finally in the fourth phase, a classification model is learnt using the ranked features. We have also performed Friedman statistical test [24] to determine the statistical difference among the proposed method and the existing methods i.e. CSP, SBCSP, FBCSP and CVSCSP.
The major contributions of this paper include:
-
(a)
The proposed method utilized relevant temporal, spectral and spatial information to distinguish motor imagery tasks.
-
(b)
Four univariate filter feature selection methods are investigated to find a reduced subset of relevant features for motor imagery tasks classification.
-
(c)
The performance of the proposed method is compared with the existing methods on two publicly available datasets. Friedman statistical test is employed to show that the proposed method statistically significantly outperformed the existing methods.
Rest of the article is organized as follows: “Related works” section includes related research works of motor imagery BCI. In “Combined variable sized subband and temporal filter based stationary common spatial patterns (CVSTSCSP)” section, we discuss the proposed method. “Experimental data and results” section illustrates the experimental setup and results. Finally, in “Conclusion and future directions” section, conclusion of the article with some future insights are discussed.
Related works
CSP is one of primary spatial filtering techniques used in the area of motor imagery BCI. However, CSP performs poorly due to problems like non-stationarity of EEG signals, artefacts generated from eye movements, electromyographic activity or any other muscular movement, irrelevant frequency filtering, etc. [2, 17]. CSP variants like common spatio-spectral pattern (CSSP) and common sparse spectral spatial pattern (CSSSP) include time delay embedding to optimize spectral filters simultaneously with the optimization of CSP filters. These methods are able to overcome some of the limitations faced by CSP. However, due to multiple time delay embedding and regularization of classifier parameters, space and time complexity of these techniques is quite high [25]. Another variant of CSP i.e. the stationary common spatial pattern (SCSP) has been proposed to reduce the effect of non-stationary characteristics of the EEG signal by introducing a penalty term in the CSP’s target function [17]. Further in this direction, to take care of non-stationary and variable nature of EEG signals, non-homogenous spatial filters (distinct frequency and time dependent spatial filters) have been used [26]. In a similar kind of research work [23, 27], spatial and spatio-spectral filters are estimated by a generalized CSP framework using an optimization constraint and specific target function for improving classification performance and reducing the instability caused by non-stationarity of EEG signals. Subject transfer based composite local temporal correlation CSP has been proposed to deal with noise and inter subject variability using the concept of local temporal based covariance matrices and composite approach based subject in the research work [9]. All these methods, performs simultaneous optimization of spatial or spectral filters within CSP optimization criterion.
On the other hand, instead of simultaneous optimization of a spectral filter within CSP, some of the other variants of CSP select significant features from multiple frequency bands to improve classification performance. Subband CSP [19] extracted spatial filters features from a non-overlapping fixed size subband frequency filter bank and used LDA score fusion for classification. SBCSP uses RFE SVM based feature selection to remove irrelevant subband features. In the research work [20], mutual information has been used as a feature selection criterion that considers the nonlinear correlation between the features from different frequency bands and the class variable. In another research work [28], sparse filter band CSP has been proposed that uses overlapping fixed sized subbands of a frequency band to optimise the CSP feature selection with the lasso estimate. In the similar research direction, spatial spectral filters are optimised in [29] with the aim of minimizing Bayesian classification error while maximizing mutual information among the frequency bands. However, all these variants of CSP, uses fixed size subband filters for feature extraction. The research work proposed by [23] have suggested the use of backtracking search optimization algorithm for relevant frequency band and time segment selection for motor imagery BCI. However, evolutionary algorithms are computationally intensive. Also, these methods require tuning of more number of parameters such as kind of selection, crossover operator, population size, fitness function to achieve optimal solution [30]. Hence, evolutionary algorithms are not suitable for real time BCI application. Further, most of the research work discussed in literature considers features based on temporal, spatial and spectral content separately or in combination of two and not all of the three simultaneously.
Combined variable sized subband and temporal filter based stationary common spatial patterns (CVSTSCSP)
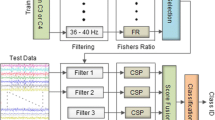
Figure 1 shows the flow diagram of the four phases used in proposed CVSTSCSP model. A brief description of each of these phases is given below:
Preprocessing
-
Data segmentation
In the proposed method, we segment the raw data into three overlapping time windows [TS1:0.5–2.5, TS2: 1.0–3.0, TS3:1.5–3.5]. The data from time window [0.0–0.5 and 3.5–4.0] has not been used to avoid the overlapping from resting data [22].
-
Generation of Frequency Band and Bandpass Filtering
In this step, we generate frequency subbands of variable size from a defined frequency range, a minimum bandwidth and a defined frequency granularity i.e. the length between the central frequencies of two contiguous bands. The generated set of frequency subbands act as a filter bank of variable size subbands. The process to generate variable sized frequency filter bank explicitly does not need information of relevant subbands to distinguish given two motor imagery tasks. As an example, various subbands generated for frequency range (7–32), a minimum bandwidth and granularity of 5 Hz can be seen in Fig. 2. These variable sized set of subbands act as overlapping subband filter bank. The data obtained from each of the time segment is bandpass filtered from this filter bank of variable sized subband filters.
Feature extraction
SCSP
To extract relevant features filtered data, stationary common spatial patterns (SCSP) [17] technique is utilized, which is a variant of CSP spatial filtering technique. In CSP, spatial filters are derived from the simultaneous diagonalization of the covariance matrices of the EEG signal data from each class that can be achieved by solving the optimization problem of the following Rayleigh criterion maximization function:
where Σ1and Σ2 are normalized average covariance matrices of class1 and class 2 respectively and W is a spatial filter matrix. However, CSP suffers from problems like presence of artifacts and stationarities within the signal. In SCSP, to minimize the effect of non-stationarity, a measure of stationarity has been used which is given by the sum of absolute differences between the projected average variance of all trials and the projected variance in each trial. The difference between normalized average covariance Σ1or Σ2 and the covariance matrix of each trial k for each class is given by:
where s is an operator for making symmetric matrix positive definite. The average difference matrix for class c (c = 1, 2) is given as:
The modified Rayleigh Criterion maximization function is given as:
where \( \mathrm{P}\left(\mathbf{W}\right)={\mathbf{W}}^{\mathrm{T}}\left({\overline{\varDelta}}_1+{\overline{\varDelta}}_2\right)\mathbf{W} \) is penalty term and α is a constant determined using method proposed in [31]. The transformed matrix Z for a given trial X, is given as:
Feature fp is computed as:
where Zp are the first and last r rows of Z.
LDA
SCSP features, extracted from each combination of TSth time segment and kth subband, are transformed using linear discriminant analyser which provides a projection matrix \( {\mathbf{W}}_{\mathbf{lda}}^{\mathbf{TS},\mathbf{k}} \) that minimizes intra class variance \( {\mathbf{S}}_{\mathrm{W}}^{\mathrm{TS},\mathrm{k}} \) and maximizes inter class variance \( {\mathbf{S}}_{\mathrm{B}}^{\mathrm{TS},\mathrm{k}} \) given by:
and
The cost function for TSth time segment and kth subband, which needs to be maximized, is given by:
where \( {\mathbf{m}}_1^{\mathrm{TS},\mathrm{k}} \) and \( {\mathbf{m}}_2^{\mathrm{TS},\mathrm{k}} \) are means of class 1 and class 2 features for TSth time segment and kth subband respectively. The score for TSth time segment and kth subband is defined as
The scores obtained from each combination of TSth time segment and kth subband are fused to form a 3*k-dimensional feature vector \( {\left[{\mathbf{s}}_1^{\mathbf{T}\mathbf{S}1},{\mathbf{s}}_2^{\mathbf{T}\mathbf{S}1}\dots {\mathbf{s}}_{\mathbf{k}}^{\mathbf{T}\mathbf{S}1},{\mathbf{s}}_1^{\mathbf{T}\mathbf{S}2},{\mathbf{s}}_2^{\mathbf{T}\mathbf{S}2}\dots {\mathbf{s}}_{\mathbf{k}}^{\mathbf{T}\mathbf{S}2},{\mathbf{s}}_1^{\mathbf{T}\mathbf{S}3},{\mathbf{s}}_2^{\mathbf{T}\mathbf{S}1}\dots {\mathbf{s}}_{\mathbf{k}}^{\mathbf{T}\mathbf{S}3}\right]}^{\mathbf{T}} \) corresponding to each trial.
Feature selection
Feature vector derived may enclose features from irrelevant time segment and subband for a given mental task. These irrelevant features may deteriorate the performance of decision model. In order to gain proper insight about the features and their relevance to a class variable, univariate feature selection techniques have been used in literature [32]. A subset of feature are selected on the basis of a selection criterion. The reduced and relevant set of features will require reduced space and less computation time for leaning a model and will also provide improved classification performance. Thus, the ranking of features is carried out in the third phase using following univariate feature selection approaches:
-
Euclidean distance: Euclidean distance [32] is a Pythagorean Theorem based simple distance measure. It measures the distance between the data points belonging to two classes. Euclidean distance between the two classes c 1 and c 2 for including the feature f is given by:
where \( {\mu}_i^{\mathrm{f}} \) is the mean of feature f for class ci. The high value of D1,2 characterizes that the two classes are highly distinguishing. It is simple and fast. However, it assumes that the samples are distributed about its mean spherically.
-
Correlation: Correlation [33] is adopted as a measure of goodness between two variables (feature f and class variable c) with the assumption that a good feature is highly correlated to class label. The linear correlation coefficient between class ci (i = 1, 2) and feature f is given by:
where \( {\mu}_i^{\mathrm{f}} \) is the mean of feature f and \( {\mu}_i^{\mathrm{c}} \) is the mean of class ci. The value of r lies between −1 and 1. Higher the magnitude of R, more relevant is that feature f.
-
Mutual information: The mutual information [34] measures the nonlinear correlation between two random variables. The mutual information I(c; f) between class ci (i = 1, 2) and feature f is given by:
where H(ci) is the entropy function for class variable c given by:
and H(ci| f) is the change in entropy value of class variable c by observing feature f is given by:
where P(ci) is the probability density function for class variable ci and P(ci | f) is the conditional probability density function. Higher the magnitude of I(ci; f), more relevant is that feature f to the class variable ci. Given an initial set of features, a subset of features that provides maximal mutual information is selected for classification.
-
Fisher discriminant ratio (FDR): FDR [35] is a ranking approach that ranks the features on the basis of following measure:
where μi and σi denote mean and variance of the ith class features, respectively. Higher value of FDR depicts the data of different classes is more separable and less scattered around their mean.
Classification
After obtaining relevant features, a decision model is built. Two well-known classifiers such as linear discriminant analysis (LDA) [36] and support vector machine (SVM) [33] are investigated in this paper.
Experimental data and results
Dataset description and parameter setting
Dataset 1: “BCI competition III dataset Iva”
Fraunhofer FIRST and Campus Benjamin Franklin of the Charite - University Medicine Berlin have provided this dataset [37]. The given dataset is composed of motor Imagery EEG signals generated during right hand and right foot motor imagination. Five healthy subjects (aa, al, av., aw and ay) were employed for acquisition of data. Each subject’s dataset consists of EEG signals of 280 trials. The signals were measured using 118 EEG channel locations from extended international 10/20 electrode montage system. During each trial, the subject was provided with a visual cue shown for 3.5 s showing which of the three motor imagery tasks, the subject needs to perform: left hand motor imagery, right hand motor imagery, and right foot motor imagery. The captured EEG data was preprocessed using a bandpass filter of 0.05–200 Hz and then fed to digitization at 1000 Hz and downsampled at 100 Hz. The resting window between two adjacent experiments was randomly taken from a time period of 1.75–2.25 s. EEG trials only for the right-hand motor imagery and right-foot motor imagery were given for competition purpose. These parameter settings are provided by the BCI competition.
Dataset 2: “BCI competition IV dataset Ia”
Berlin BCI group, Fraunhofer FIRST and Campus Benjamin Franklin of the Charité University Medicine Berlin have provided this dataset [38]. Seven healthy subjects (ds1a, ds1b, ds1c, ds1d, ds1e, ds1f and ds1g) were employed for acquisition of data. Each subject’s dataset consists of EEG signals of 200 trials. The signals were measured using 59 EEG channel locations from extended international 10/20 electrode montage system. For the period of each trial, the subject was provided with a visual cue shown for 4 s. For each subject, two classes of motor imagery were selected from the three classes left hand motor imagery, right hand motor imagery, and foot motor imagery at a time. The whole dataset is divided into two categories: Calibration Data and Evaluation Data. The captured EEG Signals were bandpass filtered between 0.05 and 200 Hz and then fed to digitization at 1000 Hz and downsampled at 100 Hz. The resting window between two adjacent experiments was randomly taken from a time period of 2–4 s. These parameter settings are provided by the BCI competition.
For experimental analysis, we have used the data from each trial that belongs to the overlapping time windows [0.5 to 2.5, 1.0 to 3.0, and 1.5 to 3.5 s] [22] after the onset of stimulus which yields a total of 200-time units per electrode in a trial or an EEG signal matrix of 118 × 200 per trial for Dataset 1 and 59 × 200 [21, 31, 39] per trial for Dataset 2. Variable size subbands is generated from a given frequency band [7–30 Hz] at a variable bandwidth (bw) = [3,4,…7 Hz] and granularity (gr) = [3,4,…7 Hz]. Thus, the smallest considered bandwidth of variable subbands is not very small thus information loss would be minimal. The time segmented data is then bandpass filtered using the different variable sized subbands filter bank. Stationary CSP in combination with LDA is then used for extracting features. SCSP penalty parameter μ = 0.1 (decided using cross-validation) is used for all the experiments on both datasets. In literature [18], it is shown that r= 1 or r = 2 is a good choice and adding more number of spatial patterns does not enhance the classification performance. Therefore, in this research work, the number of spatial patterns has been fixed to r = 1 [19,20,21]. Hence for each time segment and variable size band combination, we obtain two features for each trial and LDA analysis converts the obtained two features from each combination of time segment and subband into one feature. Univariate feature selection methods are then used to rank these features. To achieve the best performance of learning machine, grid search is employed to obtain SVM regularization parameter C and the Gaussian kernel parameter σ which varied from 1 to 500 and 1 to 100 respectively. The optimal value so obtained were C = 100 and σ = 10 based on experimentation.
The performance of the CKSSCSP is compared with existing methods (CSP, SBCSP, FBCSP and CVSCSP) in terms of average classification error. The classification error is reported as average of 10 runs of 10-fold cross-validation classification error for each subject. In our experiment, for comparison with existing methods, we have performed experiments on the parameters that were suggested in the existing works [19,20,21]. Therefore, in this work, a fixed size bw = 4 Hz is considered for SBCSP, FBCSP and for CVSCSP bw = 4 Hz, gr = 4 Hz and Euclidean distance based feature selection are used for performance evaluation. Different values of bw and gr ranging from 3 to 7 Hz have been used for evaluating the variation in the performance of each subject using proposed method CVSTSCSP.
Results and discussion
Figures 3, 4, 5 and 6 show variations in classification error of the proposed method CVSTSCSP with the choice of single time segments (TS1:0.5–2.5, TS2: 1.0–3.0, TS3:1.5–3.5), and all three segments (ALL_TS)) of Dataset 1 and Dataset 2 for SVM and LDA classifiers respectively using different univariate feature selection methods. The classification error is reported in terms of average classification error of 10 runs of 10-fold cross-validation for all subjects. The following can be noted from Figs. 3 and 4 for Dataset 1:
-
Using correlation based feature selection, there is an overall decrease of 23.04%, 30.9% and 24.68% in average classification error using LDA classifier and a decrease of 10.65%, 24.29% and 24.77% in average classification error using SVM classifier with the use of all relevant features from the three time segments ALL_TS in CVSTSCSP as compared to relevant features from single time segment TS1, TS2 and TS3 in CVSTSCSP respectively.
-
Using FDR based feature selection, there is an overall decrease of 25.12%, 32.25% and 24.98% in average classification error using LDA classifier and a decrease of 9.33%, 23.65% and 24.45% in average classification error using SVM classifier with the use of all relevant features from the three time segments ALL_TS in CVSTSCSP as compared to relevant features from single time segment TS1, TS2 and TS3 in CVSTSCSP respectively.
-
Using Euclidean based feature selection, there is an overall decrease of 6%, 3.2% and 12.68% in average classification error using LDA classifier and a decrease of 16.89%, 16.53% and 35.38% in average classification error using SVM classifier with the use of all relevant features from the three time segments ALL_TS in CVSTSCSP as compared to relevant features from single time segment TS1, TS2 and TS3 in CVSTSCSP respectively.
-
Using mutual information based feature selection, there is an overall decrease of 35.7%, 32.5% and 41.3% in average classification error using LDA classifier and a decrease of 17.09%, 13.98% and 41.04% in average classification error using SVM classifier with the use of all relevant features from the three time segments ALL_TS in CVSTSCSP as compared to relevant features from single time segment TS1, TS2 and TS3 in CVSTSCSP respectively.
-
The combination of CVSTSCSP with mutual information based feature selection performs best and the combination of CVSTSCSP with Euclidean distance based feature selection performs worst among all combinations of feature selection with CVSTSCSP with SVM classifier.
-
The combination of CVSTSCSP with mutual information based feature selection performs best and the combination of CVSTSCSP with correlation based feature selection performs worst among all combinations of feature selection with CVSTSCSP with LDA classifier.
-
On an average, minimum average classification error of 5.1% and 4.48% is obtained using mutual information based feature selection for Dataset 1 with SVM and LDA classifier respectively. Finally, we can also observe from Figs. 3 and 4 that classification error has reduced significantly with the use of all relevant features from the three time segments ALL_TS as compared to relevant features from single time segment TS1, TS2 and TS3 in the proposed CVSTSCSP method with SVM as well as LDA classifier.
Following deductions can be made from Figs. 5 and 6 for Dataset 2:
-
Using correlation based feature selection, there is an overall decrease of 39.93%, 17.28% and 33.18% in average classification error using LDA classifier and a decrease of 43.75%, 32.91% and 41.12% in average classification error using SVM classifier with the use of all relevant features from the three time segments ALL_TS in CVSTSCSP as compared to relevant features from single time segment TS1, TS2 and TS3 in CVSTSCSP respectively.
-
Using FDR based feature selection, there is an overall decrease of 39.98%, 17.42% and 33.20% in average classification error using LDA classifier and a decrease of 43.69%, 32.84% and 41.05% in average classification error using SVM classifier with the use of all relevant features from the three time segments ALL_TS in CVSTSCSP as compared to relevant features from single time segment TS1, TS2 and TS3 in CVSTSCSP respectively.
-
Using Euclidean based feature selection, there is an overall decrease of 29.38%, 15.62% and 23.22% in average classification error using LDA classifier and a decrease of 38.07%, 28.25% and 31.31% in average classification error using SVM classifier with the use of all relevant features from the three time segments ALL_TS in CVSTSCSP as compared to relevant features from single time segment TS1, TS2 and TS3 in CVSTSCSP respectively.
-
Using mutual information based feature selection, there is an overall decrease of 38.98%, 14.94% and 31.93% in average classification error using LDA classifier and a decrease of 43.42%, 33.48% and 41.21% in average classification error using SVM classifier with the use of all relevant features from the three time segments ALL_TS in CVSTSCSP as compared to relevant features from single time segment TS1, TS2 and TS3 in CVSTSCSP respectively.
-
The combination of CVSTSCSP with mutual information based feature selection performs best and the combination of CVSTSCSP with Euclidean distance based feature selection performs worst among all combinations of feature selection with CVSTSCSP with SVM classifier.
-
The combination of CVSTSCSP with mutual information based feature selection performs best and the combination of CVSTSCSP with correlation based feature selection performs worst among all combinations of feature selection with CVSTSCSP with LDA classifier.
-
On an average, a minimum average classification error of 6.46% is obtained using correlation based feature selection with SVM classifier and a minimum average classification error of 7.68% is obtained using FDR based feature selection with LDA classifier for Dataset 2.
-
Finally, we can also observe from Figs. 5 and 6 that classification error has reduced significantly with the use of all relevant features from the three time segments ALL_TS as compared to relevant features from single time segment TS1, TS2 and TS3 in the proposed CVSTSCSP method with SVM as well as LDA classifier
Tables 1 and 2 show the comparison of average classification error of all the existing methods with the proposed approach CVSTSCSP for dataset1 with LDA and SVM classifiers respectively. From Tables 1 and 2, the following observations can be deduced for Dataset 1:
-
The proposed method CVSTSCSP performs best among all methods and achieves a minimum classification error of 0.04 and 0.05 with LDA and SVM classifier respectively.
-
There is an overall decrease of 80.72% and 82.2% in classification error with the use of CVSTSCSP in comparison to CSP with LDA and SVM classifier respectively.
-
An overall decrease of 82.0% and 79.82% in classification error can be observed using the proposed method CVSTSCSP in comparison to SBCSP with LDA and SVM respectively.
-
A decrease of 82.3% and 84.2% classification error has been achieved with CVSTSCSP in comparison to FBCSP with LDA and SVM classifier respectively.
-
A deduction in classification error of 86.2% and 89.1% has also been obtained with the proposed method CVSTSCSP in comparison to CVSCSP method.
-
An average decrease of 81.1%, 80.9%, 83.3% and 87.7% can be observed using the proposed method CVSTSCSP in comparison to CSP, SBCSP, FBCSP and CVSCSP respectively over all classifiers and all subjects of Dataset 1.
Tables 3 and 4 shows the comparison of average classification error of all the existing methods with the proposed approach for Dataset 2 with LDA and SVM classifiers respectively. From Tables 3 and 4, following observations can be deduced for Dataset 2 as above:
-
The proposed method CVSTSCSP performs best among all methods and achieves a minimum classification error of 0.08 and 0.07 with LDA and SVM classifier respectively.
-
There is an overall decrease of 79.8% and 76.7% in classification error with the use of CVSTSCSP as compared to CSP with LDA and SVM classifier respectively.
-
An overall decrease of 78.7% and 72.1% in classification error can be observed using the proposed method as compared to SBCSP with LDA and SVM respectively.
-
A decrease of 82.2% and 79.4% classification error has been achieved with CVSTSCSP in comparison to FBCSP with LDA and SVM classifier respectively.
-
A deduction in classification error of 85.2% and 84.38% have also been obtained while comparing the proposed method and CVSCSP method.
-
An average decrease of 78.5%, 75.4%, 80.8% and 84.80% in classification error can be observed using the proposed method CVSTSCSP in comparison to CSP, SBCSP, FBCSP and CVSCSP respectively over all classifiers and all subjects of Dataset 2.
An average decrease of 79.7%, 78.20%, 82.1% and 86.26% in classification error over both datasets and both classifiers can be observed using the proposed method CVSTSCSP in comparison to CSP, SBCSP, FBCSP and CVSCSP respectively.
Figures 7 and 8 show the variations in classification error with different combinations of bandwidth (bw) and granularilty (gr) values for different subjects of Dataset 1 and Dataset 2 respectively. We can observe from Figs. 7 and 8 that the classification error varies with the choice of bandwidth (bw) and granularilty (gr). Also, the minimum classification error is achieved with different combination of bw and gr for different subjects. It can also be noted that the classification error is more sensitive to the choice of bw in comparison to gr. Further, it can be observed that larger values of gr and bw leads to a degraded performance as the number of bands generated are less in number and may not consists of the relevant subset of subbands to a particular subject.
To find the statistical difference among all the experimental methods, Friedman statistical tests [20] have been conducted in this study. The null hypothesis assumes that the performance of all the methods is equivalent in terms of classification error. Table 5 shows the statistical ranking of all the method obtained using Friedman’s statistical test. It can be observed from Table 5 that the proposed method CVSTSCSP in combination with mutual information based feature selection (mi-CVSTSCSP–All_TS) performs the best, which has achieved the least rank value of 3.167. The p value calculated using Iman and Davenport statistic [20] is 3.06E-37 which confirms the significant difference among the all the methods used in our experimental study. Thus, we can reject the null hypothesis and state that the proposed method is statistically significant.
To compare all other methods with the best ranked method i.e. control method (mi-CVSTSCSP–All_TS), p values are computed using a defined post hoc methods (Hommel, Holm and Hochberg methods) [32]. Table 6 shows the p values obtained for defined post hoc methods. The bold value highlights the significant difference of the control method (mi-CVSTSCSP–All_TS) with all other methods at a significance level of 0.05 using the post hoc methods.
Conclusion and future directions
Many Feature extraction techniques have been used in the area of BCI for recognition of motor imagery tasks. CSP is one of the popular spatial feature extraction method used in the area of motor imagery EEG classification. The performance of CSP is highly dependent on subject specific characteristics like frequency band, relevant time segment within a trial, spatial filters, and presence of artifacts in the EEG signal.
In this paper, we proposed a four-phase method CVSTSCSP to determine relevant features from a set of spectral, temporal and spatial features to reduce the classification error to distinguish motor imagery tasks. In order to determine relevant temporal information, the EEG signal is segmented into three overlapping time segments. Further, to choose the relevant spectral features, we have used variable sized subband filter bank. To reduce the effect of artifacts and non-stationarity, SCSP is used for feature extraction. In order to select a reduced subset of relevant subset of features from high-dimensional feature vector, we have used univariate feature selection method. We have investigated four univariate feature selection methods such as Euclidean distance, correlation, mutual information and Fisher discriminant ratio. Two well-known classifiers LDA and SVM are used to the build decision model. It is observed that with the use of relevant temporal information in the proposed CVSTSCSP method, the performance improves in terms of the classification error in comparison to the CVSCSP method, which consider the whole signal. The combination of CVSTSCSP with mutual information based feature selection achieves minimum classification error for Dataset 1 and comparable classification error for Dataset 2 among all combinations of CVSTSCSP with different feature selection method. It is also noted that the classification error is more sensitive to the choice of bandwidth (bw) in comparison to granularilty (gr). Experimental results demonstrate that the proposed method CVSTSCSP outperforms the existing methods such as CSP, SBCSP, FBCSP and CVSCSP in terms of classification error. Friedman statistical test has been performed to confirm the significant difference among the all the methods used in our experimental study.
For evaluation of the proposed method, we have conducted all the experiments on two class motor imagery EEG data only. In future, we will extend the proposed method for multiclass classification. Univariate feature ranking methods have been used in this study. Although these methods are simple, efficient to implement and select relevant features. But these methods ignore the correlation among the subset of relevant features, which may degrade the performance. Multivariate feature selection methods have been suggested in research work [33] which provide relevant and non-redundant subset of features. We will utilize multivariate feature selection methods in our future works. The proposed method uses static selection of subject specific time segment. In future work, we will extend our research work for automatic selection of subject specific time segment.
References
Bashashati, A., Fatourechi, M., Ward, R. K., and Birch, G. E., A survey of signal processing algorithms in brain–computer interfaces based on electrical brain signals. J. Neural Eng. 4(2):R32–R57, 2007.
Dornhege, G. ed., Toward Brain-computer Interfacing. MIT Press, 2007.
Nordin, N., Xie, S. Q., and Wünsche, B., Assessment of movement quality in robot-assisted upper limb rehabilitation after stroke: a review. J. Neuroengineering Rehabil. 11(1):137, 2014.
Li, Y., Pan J., He, Y., Wang, F., Laureys, S., Xie, Q., and Yu, R., Detecting number processing and mental calculation in patients with disorders of consciousness using a hybrid brain-computer interface system. BMC Neurol. 15(1):259, 2015.
Kasahara, T., Terasaki, K., Ogawa, Y., Ushiba, J., Aramaki, H., and Masakado, Y., The correlation between motor impairments and event-related desynchronization during motor imagery in ALS patients. BMC Neurosci. 13(1):66, 2012.
Wolpaw, J. R., and Wolpaw, E. W., Brain-computer interfaces: principles and practice. OUP: USA, 2012.
Nicolas-Alonso, L. F., and Gomez-Gil, J., Brain Computer Interfaces, a Review. Sensors. 12(12):1211–1279, 2012.
Yi, W., Qiu, S., Wang, K., Qi, H., He, F., Zhou, P., Zhang, L., and Ming, D., EEG oscillatory patterns and classification of sequential compound limb motor imagery. J. NeuroEngineering Rehabil. 13(1):259, 2016.
Hatamikia, S., and Nasrabadi, A. M., Subject transfer BCI based on Composite Local Temporal Correlation Common Spatial Pattern. Comput. Biol. Med. 64:1–11, 2015.
Yi, W., Qiu, S., Qi, H., Zhang, L., Wan, B., and Ming, D., EEG feature comparison and classification of simple and compound limb motor imagery. J. Neuroengineering Rehabil. 10(1):106, 2013.
Wolpaw, J. R., Birbaumer, N., McFarland, D. J., Pfurtscheller, G., and Vaughan, T. M., Brain–computer interfaces for communication and control. Clin. Neurophysiol. 113(6):767–791, 2002.
Pfurtscheller, G., and Neuper, C., Motor imagery and direct brain-computer communication. Proc. IEEE. 89(7):1123–1134, 2001.
Ramoser, H., Muller-Gerking, J., and Pfurtscheller, G., Optimal spatial filtering of single trial EEG during imagined hand movement. IEEE Trans. Rehabil. Eng. 8(4):441–446, 2000.
Müller-Gerking, J., Pfurtscheller, G., and Flyvbjerg, H., Designing optimal spatial filters for single-trial EEG classification in a movement task. Clin. Neurophysiol. 110(5):787–798, 1999.
Lu, H., Eng, H.-L., Guan, C., Plataniotis, K. N., and Venetsanopoulos, A. N., Regularized Common Spatial Pattern With Aggregation for EEG Classification in Small-Sample Setting. IEEE Trans. Biomed. Eng. 57(12):2936–2946, 2010.
Grosse-Wentrup, M., Schölkopf, B., and Hill, J., Causal influence of gamma oscillations on the sensorimotor rhythm. NeuroImage. 56(2):837–842, 2011.
Samek, W., Vidaurre, C., Müller, K.-R., and Kawanabe, M., Stationary common spatial patterns for brain–computer interfacing. J. Neural Eng. 9(2):026013, 2012.
McFarland, D. J., and Wolpaw, J. R., Brain-computer interfaces for communication and control. Commun. ACM 54(5):60, 2011.
Novi, Q., Guan, C., Dat, T. H., and Xue, P., Sub-band common spatial pattern (SBCSP) for brain-computer interface. Neural Engineering, 2007. CNE’07. 3rd International IEEE/EMBS Conference on. pp. 204–207. IEEE, 2007.
Ang, K. K., Chin, Z. Y., Zhang, H., and Guan, C., Filter bank common spatial pattern (FBCSP) in brain-computer interface. Neural Networks, 2008. IJCNN 2008. (IEEE World Congress on Computational Intelligence). IEEE International Joint Conference on. pp. 2390–2397. IEEE, 2008.
Kirar, J. S., and Agrawal, R. K., Optimal Spatio-spectral Variable Size Subbands Filter for Motor Imagery Brain Computer Interface. Procedia Comput. Sci. 84:14–21, 2016.
Ang, K. K., Chin, Z. Y., Zhang, H., and Guan, C., Mutual information-based selection of optimal spatial–temporal patterns for single-trial EEG-based BCIs. Brain Decod. 45(6):2137–2144, 2012.
Wei, Z., and Wei, Q., The backtracking search optimization algorithm for frequency band and time segment selection in motor imagery-based brain–computer interfaces. J. Integr. Neurosci. 15(03):347–364, 2016.
Iman, R. L., and Davenport, J. M., Approximations of the critical region of the fbietkan statistic. Commun. Stat.-Theory Methods. 9(6):571–595, 1980.
Liu, G., Huang, G., Meng, J., and Zhu, X., A frequency-weighted method combined with Common Spatial Patterns for electroencephalogram classification in brain–computer interface. Biomed. Signal Process. Control. 5(2):174–180, 2010.
Kam, T.-E., Suk, H.-I., and Lee, S.-W., Non-homogeneous spatial filter optimization for ElectroEncephaloGram (EEG)-based motor imagery classification. Neurocomputing. 108:58–68, 2013.
Fattahi, D., Nasihatkon, B., and Boostani, R., A general framework to estimate spatial and spatio-spectral filters for EEG signal classification. Neurocomputing. 119:165–174, 2013.
Zhang, Y., Zhou, G., Jin, J., Wang, X., and Cichocki, A., Optimizing spatial patterns with sparse filter bands for motor-imagery based brain–computer interface. J. Neurosci. Methods. 255:85–91, 2015.
Meng, J., Yao, L., Sheng, X., Zhang, D., and Zhu, X., Simultaneously Optimizing Spatial Spectral Features Based on Mutual Information for EEG Classification. IEEE Trans. Biomed. Eng. 62(1):227–240, 2015.
Khushaba, R. N., Al-Ani, A., and Al-Jumaily, A., Differential evolution based feature subset selection. Pattern Recognition, 2008. ICPR 2008. 19th International Conference on. pp. 1–4. IEEE, 2008.
Kirar, J. S., and Agrawal, R. K., Composite kernel support vector machine based performance enhancement of brain computer interface in conjunction with spatial filter. Biomed. Signal Process. Control. 33(Supplement C):151–160, 2017.
Guyon, I., and Elisseeff, A., An introduction to variable and feature selection. J. Mach. Learn. Res. 3(Mar):1157–1182, 2003.
Guyon, I., Weston, J., Barnhill, S., and Vapnik, V., Gene selection for cancer classification using support vector machines. Mach. Learn. 46(1):389–422, 2002.
Battiti, R., Using mutual information for selecting features in supervised neural net learning. IEEE Trans. Neural Netw. 5(4):537–550, 1994.
Webb, A. R., Statistical pattern recognition. John Wiley & Sons, 2003.
Bishop, C. M., Pattern recognition and machine learning. Springer, 2006.
Dornhege, G., Blankertz, B., Curio, G., and Muller, K.-R., Boosting bit rates in noninvasive EEG single-trial classifications by feature combination and multiclass paradigms. IEEE Trans. Biomed. Eng. 51(6):993–1002, 2004.
Blankertz, B., Dornhege, G., Krauledat, M., Müller, K.-R., and Curio, G., The non-invasive Berlin brain–computer interface: fast acquisition of effective performance in untrained subjects. NeuroImage. 37(2):539–550, 2007.
Kirar, J. S., Choudhary, A., and Agrawal, R. K., Selection of Relevant Electrodes Based on Temporal Similarity for Classification of Motor Imagery Tasks. In: Shankar, B. U., Ghosh, K., Mandal, D. P., Ray, S. S., Zhang, D., and Pal, S. K., (Eds.), Pattern Recognition and Machine Intelligence: 7th International Conference, PReMI 2017, Kolkata, India, December 5–8, 2017, Proceedings. Cham: Springer International Publishing, 2017, 96–102.
Funding
This study was funded by University Grant Commission, India.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author(s) declare that they have no conflict of interest.
Additional information
This article is part of the Topical Collection on Image & Signal Processing
Rights and permissions
About this article
Cite this article
Kirar, J.S., Agrawal, R.K. Relevant Feature Selection from a Combination of Spectral-Temporal and Spatial Features for Classification of Motor Imagery EEG. J Med Syst 42, 78 (2018). https://doi.org/10.1007/s10916-018-0931-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-018-0931-8